Locally epistatic genomic relationship matrices for genomic association and prediction
- PMID: 25614606
- PMCID: PMC4349077
- DOI: 10.1534/genetics.114.173658
Locally epistatic genomic relationship matrices for genomic association and prediction
Abstract
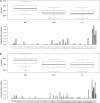
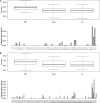
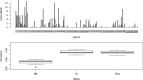
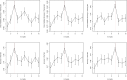
In plant and animal breeding studies a distinction is made between the genetic value (additive plus epistatic genetic effects) and the breeding value (additive genetic effects) of an individual since it is expected that some of the epistatic genetic effects will be lost due to recombination. In this article, we argue that the breeder can take advantage of the epistatic marker effects in regions of low recombination. The models introduced here aim to estimate local epistatic line heritability by using genetic map information and combining local additive and epistatic effects. To this end, we have used semiparametric mixed models with multiple local genomic relationship matrices with hierarchical designs. Elastic-net postprocessing was used to introduce sparsity. Our models produce good predictive performance along with useful explanatory information.
Keywords: GenPred; epistasis; genomic selection; mixed models; shared data resource.
Copyright © 2015 by the Genetics Society of America.
Figures







References
-
- Akdemir, D., and O. U. Godfrey, 2014 EMMREML: Fitting Mixed Models with Known Covariance Structures R Package Version 2.0. Available at: http://CRAN.R-project.org/package=EMMREML.
-
- Amemiya T., 1977. A note on a heteroscedastic model. J. Econom. 6(3): 365–370.
-
- Bach, F. R., G. R. Lanckriet, and M. I. Jordan, 2004 Multiple kernel learning, conic duality, and the SMO algorithm, p. 6 in Proceedings of the Twenty-First International Conference on Machine Learning. ACM., New York, NY DOI: 10.1145/1015330.1015424.
-
- Blanchard G., Geman D., 2005. Hierarchical testing designs for pattern recognition. Ann. Stat. 33: 1155–1202.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

