Biased signaling at chemokine receptors
- PMID: 25614627
- PMCID: PMC4392259
- DOI: 10.1074/jbc.M114.596098
Biased signaling at chemokine receptors
Abstract
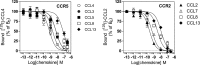
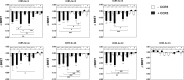
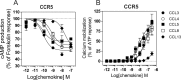
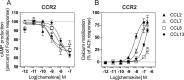
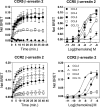
The ability of G protein-coupled receptors (GPCRs) to activate selective signaling pathways according to the conformation stabilized by bound ligands (signaling bias) is a challenging concept in the GPCR field. Signaling bias has been documented for several GPCRs, including chemokine receptors. However, most of these studies examined the global signaling bias between G protein- and arrestin-dependent pathways, leaving unaddressed the potential bias between particular G protein subtypes. Here, we investigated the coupling selectivity of chemokine receptors CCR2, CCR5, and CCR7 in response to various ligands with G protein subtypes by using bioluminescence resonance energy transfer biosensors monitoring directly the activation of G proteins. We also compared data obtained with the G protein biosensors with those obtained with other functional readouts, such as β-arrestin-2 recruitment, cAMP accumulation, and calcium mobilization assays. We showed that the binding of chemokines to CCR2, CCR5, and CCR7 activated the three Gαi subtypes (Gαi1, Gαi2, and Gαi3) and the two Gαo isoforms (Gαoa and Gαob) with potencies that generally correlate to their binding affinities. In addition, we showed that the binding of chemokines to CCR5 and CCR2 also activated Gα12, but not Gα13. For each receptor, we showed that the relative potency of various agonist chemokines was not identical in all assays, supporting the notion that signaling bias exists at chemokine receptors.
Keywords: Bioluminescence Resonance Energy Transfer (BRET); Biosensor; Chemokine; G Protein-coupled Receptor (GPCR); Signaling.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures





References
-
- Lagerström M. C., Schiöth H. B. (2008) Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug. Discov. 7, 339–357 - PubMed
-
- Strange P. G. (2008) Signaling mechanisms of GPCR ligands. Curr. Opin. Drug. Discov. Devel. 11, 196–202 - PubMed
-
- Gether U. (2000) Uncovering molecular mechanisms involved in activation of G protein-coupled receptors. Endocr. Rev. 21, 90–113 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

