The systematic study of circulating tumor cell isolation using lithographic microfilters
- PMID: 25614802
- PMCID: PMC4299665
- DOI: 10.1039/C3RA46839A
The systematic study of circulating tumor cell isolation using lithographic microfilters
Abstract
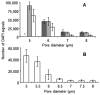
Circulating tumor cells (CTCs) disseminated into peripheral blood from a primary, or metastatic, tumor can be used for early detection, diagnosis and monitoring of solid malignancies. CTC isolation by size exclusion techniques have long interested researchers as a simple broad based approach, which is methodologically diverse for use in both genomic and protein detection platforms. Though a variety of these microfiltration systems are employed academically and commercially, the limited ability to easily alter microfilter designs has hindered the optimization for CTC capture. To overcome this problem, we studied a unique photo-definable material with a scalable and mass producible photolithographic fabrication method. We use this fabrication method to systematically study and optimize the parameters necessary for CTC isolation using a microfiltration approach, followed by a comparison to a "standard" filtration membrane. We demonstrate that properly designed microfilters can capture MCF-7 cancer cells at rate of 98 ± 2% if they consist of uniform patterned distributions, ≥160 000 pores, and 7 μm pore diameters.
Figures




References
-
- Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Clin. Cancer Res. 2004;10:6897–6904. - PubMed
-
- Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H, Terstappen LW, Meropol NJ. J. Clin. Oncol. 2008;26:3213–3221. - PubMed
-
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. N. Engl. J. Med. 2004;351:781–791. - PubMed
-
- Hou JM, Krebs MG, Lancashire L, Sloane R, Backen A, Swain RK, Priest LJ, Greystoke A, Zhou C, Morris K, Ward T, Blackhall FH, Dive C. J. Clin. Oncol. 2012;30:525–532. - PubMed
-
- Lianidou ES, Markou A. Clin. Chem. Lab. Med. 2011;49:1579–1590. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
