The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy
- PMID: 25614874
- PMCID: PMC4303222
- DOI: 10.1002/mgg3.106
The allelic spectrum of Charcot-Marie-Tooth disease in over 17,000 individuals with neuropathy
Abstract
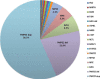
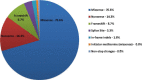
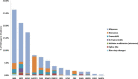
We report the frequency, positive rate, and type of mutations in 14 genes (PMP22, GJB1, MPZ, MFN2, SH3TC2, GDAP1, NEFL, LITAF, GARS, HSPB1, FIG4, EGR2, PRX, and RAB7A) associated with Charcot-Marie-Tooth disease (CMT) in a cohort of 17,880 individuals referred to a commercial genetic testing laboratory. Deidentified results from sequencing assays and multiplex ligation-dependent probe amplification (MLPA) were analyzed including 100,102 Sanger sequencing, 2338 next-generation sequencing (NGS), and 21,990 MLPA assays. Genetic abnormalities were identified in 18.5% (n = 3312) of all individuals. Testing by Sanger and MLPA (n = 3216) showed that duplications (dup) (56.7%) or deletions (del) (21.9%) in the PMP22 gene accounted for the majority of positive findings followed by mutations in the GJB1 (6.7%), MPZ (5.3%), and MFN2 (4.3%) genes. GJB1 del and mutations in the remaining genes explained 5.3% of the abnormalities. Pathogenic mutations were distributed as follows: missense (70.6%), nonsense (14.3%), frameshift (8.7%), splicing (3.3%), in-frame deletions/insertions (1.8%), initiator methionine mutations (0.8%), and nonstop changes (0.5%). Mutation frequencies, positive rates, and the types of mutations were similar between tests performed by either Sanger (n = 17,377) or NGS (n = 503). Among patients with a positive genetic finding in a CMT-related gene, 94.9% were positive in one of four genes (PMP22, GJB1, MPZ, or MFN2).
Keywords: Charcot–Marie–Tooth disease; genetic testing; high-throughput nucleotide sequencing; molecular epidemiology; peripheral neuropathy.
Figures
References
-
- Aronesty E. ea-utils: command-line tools for processing biological sequencing data. 2011. Available at http://code.google.com/p/ea-utils (accessed 8 January 2014)
-
- Boerkoel CFTH, Garcia CA, Olney RK, Johnson J, Berry K, Russo P, et al. Charcot-Marie-Tooth disease and related neuropathies: mutation distribution and genotype-phenotype correlation. Ann. Neurol. 2002;51:190–201. - PubMed
-
- CDC. 2007. Center for disease control office of public health genomics, ACCE Project.
-
- De Jonghe P, Timmerman V, Nelis E, Martin JJ. Van Broeckhoven C. Charcot-Marie-Tooth disease and related peripheral neuropathies. J. Peripher. Nerv. Syst. 1997;2:370–387. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

