The Arabidopsis SWI2/SNF2 chromatin Remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP
- PMID: 25615622
- PMCID: PMC4304717
- DOI: 10.1371/journal.pgen.1004944
The Arabidopsis SWI2/SNF2 chromatin Remodeler BRAHMA regulates polycomb function during vegetative development and directly activates the flowering repressor gene SVP
Abstract
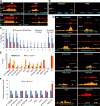
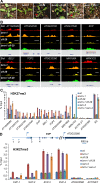
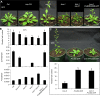
The chromatin remodeler BRAHMA (BRM) is a Trithorax Group (TrxG) protein that antagonizes the functions of Polycomb Group (PcG) proteins in fly and mammals. Recent studies also implicate such a role for Arabidopsis (Arabidopsis thaliana) BRM but the molecular mechanisms underlying the antagonism are unclear. To understand the interplay between BRM and PcG during plant development, we performed a genome-wide analysis of trimethylated histone H3 lysine 27 (H3K27me3) in brm mutant seedlings by chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq). Increased H3K27me3 deposition at several hundred genes was observed in brm mutants and this increase was partially supressed by removal of the H3K27 methyltransferase CURLY LEAF (CLF) or SWINGER (SWN). ChIP experiments demonstrated that BRM directly binds to a subset of the genes and prevents the inappropriate association and/or activity of PcG proteins at these loci. Together, these results indicate a crucial role of BRM in restricting the inappropriate activity of PcG during plant development. The key flowering repressor gene SHORT VEGETATIVE PHASE (SVP) is such a BRM target. In brm mutants, elevated PcG occupancy at SVP accompanies a dramatic increase in H3K27me3 levels at this locus and a concomitant reduction of SVP expression. Further, our gain- and loss-of-function genetic evidence establishes that BRM controls flowering time by directly activating SVP expression. This work reveals a genome-wide functional interplay between BRM and PcG and provides new insights into the impacts of these proteins in plant growth and development.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Genome-wide occupancy of Arabidopsis SWI/SNF chromatin remodeler SPLAYED provides insights into its interplay with its close homolog BRAHMA and Polycomb proteins.Plant J. 2021 Apr;106(1):200-213. doi: 10.1111/tpj.15159. Epub 2021 Mar 17. Plant J. 2021. PMID: 33432631
-
Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture.PLoS Genet. 2015 Mar 30;11(3):e1005125. doi: 10.1371/journal.pgen.1005125. eCollection 2015 Mar. PLoS Genet. 2015. PMID: 25822547 Free PMC article.
-
Kicking against the PRCs - A Domesticated Transposase Antagonises Silencing Mediated by Polycomb Group Proteins and Is an Accessory Component of Polycomb Repressive Complex 2.PLoS Genet. 2015 Dec 7;11(12):e1005660. doi: 10.1371/journal.pgen.1005660. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26642436 Free PMC article.
-
The complexity of PRC2 catalysts CLF and SWN in plants.Biochem Soc Trans. 2020 Dec 18;48(6):2779-2789. doi: 10.1042/BST20200660. Biochem Soc Trans. 2020. PMID: 33170267 Review.
-
An insight into understanding the coupling between homologous recombination mediated DNA repair and chromatin remodeling mechanisms in plant genome: an update.Cell Cycle. 2021 Sep;20(18):1760-1784. doi: 10.1080/15384101.2021.1966584. Epub 2021 Aug 26. Cell Cycle. 2021. PMID: 34437813 Free PMC article. Review.
Cited by
-
Photoperiodic Regulation of Florigen Function in Arabidopsis thaliana.Arabidopsis Book. 2015 Jun 24;13:e0178. doi: 10.1199/tab.0178. eCollection 2015. Arabidopsis Book. 2015. PMID: 26157354 Free PMC article.
-
Chromatin remodeling complexes regulate genome architecture in Arabidopsis.Plant Cell. 2022 Jul 4;34(7):2638-2651. doi: 10.1093/plcell/koac117. Plant Cell. 2022. PMID: 35445713 Free PMC article.
-
Linking transcriptional silencing with chromatin remodeling, folding, and positioning in the nucleus.Curr Opin Plant Biol. 2022 Oct;69:102261. doi: 10.1016/j.pbi.2022.102261. Epub 2022 Jul 13. Curr Opin Plant Biol. 2022. PMID: 35841650 Free PMC article. Review.
-
Comprehensive Survey of ChIP-Seq Datasets to Identify Candidate Iron Homeostasis Genes Regulated by Chromatin Modifications.Methods Mol Biol. 2023;2665:95-111. doi: 10.1007/978-1-0716-3183-6_9. Methods Mol Biol. 2023. PMID: 37166596
-
Spotlight on Plant Bromodomain Proteins.Biology (Basel). 2023 Aug 2;12(8):1076. doi: 10.3390/biology12081076. Biology (Basel). 2023. PMID: 37626962 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

