Effect of interior loop length on the thermal stability and pK(a) of i-motif DNA
- PMID: 25619229
- PMCID: PMC4630671
- DOI: 10.1021/bi5014722
Effect of interior loop length on the thermal stability and pK(a) of i-motif DNA
Abstract
The four-stranded i-motif (iM) conformation of cytosine-rich DNA is important in a wide variety of biochemical systems ranging from its use in nanomaterials to a potential role in oncogene regulation. An iM is stabilized by acidic pH that allows hemiprotonated cytidines to form a C·C(+) base pair. Fundamental studies that aim to understand how the lengths of loops connecting the protonated C·C(+) pairs affect intramolecular iM physical properties are described here. We characterized both the thermal stability and the pK(a) of intramolecular iMs with differing loop lengths, in both dilute solutions and solutions containing molecular crowding agents. Our results showed that intramolecular iMs with longer central loops form at pHs and temperatures higher than those of iMs with longer outer loops. Our studies also showed that increases in thermal stability of iMs when molecular crowding agents are present are dependent on the loop that is lengthened. However, the increase in pK(a) for iMs when molecular crowding agents are present is insensitive to loop length. Importantly, we also determined the proton activity of solutions containing high concentrations of molecular crowding agents to ascertain whether the increase in pK(a) of an iM is caused by alteration of this activity in buffered solutions. We determined that crowding agents alone increase the apparent pK(a) of a number of small molecules as well as iMs but that increases to iM pK(a) were greater than that expected from a shift in proton activity.
Figures

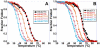
Similar articles
-
Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA.Biochemistry. 2014 Mar 18;53(10):1586-94. doi: 10.1021/bi401523b. Epub 2014 Mar 6. Biochemistry. 2014. PMID: 24564458 Free PMC article.
-
Systematic investigation of sequence requirements for DNA i-motif formation.Nucleic Acids Res. 2019 Mar 18;47(5):2177-2189. doi: 10.1093/nar/gkz046. Nucleic Acids Res. 2019. PMID: 30715498 Free PMC article.
-
Unraveling the 4n - 1 rule for DNA i-motif stability: base pairs vs. loop lengths.Org Biomol Chem. 2018 Jun 20;16(24):4537-4546. doi: 10.1039/c8ob01198b. Org Biomol Chem. 2018. PMID: 29873385
-
Driving forces for nucleic acid pK(a) shifting in an A(+).C wobble: effects of helix position, temperature, and ionic strength.Biochemistry. 2010 Apr 20;49(15):3225-36. doi: 10.1021/bi901920g. Biochemistry. 2010. PMID: 20337429 Review.
-
i-Motif DNA: structure, stability and targeting with ligands.Bioorg Med Chem. 2014 Aug 15;22(16):4407-18. doi: 10.1016/j.bmc.2014.05.047. Epub 2014 Jun 4. Bioorg Med Chem. 2014. PMID: 24957878 Review.
Cited by
-
Structure, Properties, and Biological Relevance of the DNA and RNA G-Quadruplexes: Overview 50 Years after Their Discovery.Biochemistry (Mosc). 2016 Dec;81(13):1602-1649. doi: 10.1134/S0006297916130034. Biochemistry (Mosc). 2016. PMID: 28260487 Free PMC article.
-
A single molecule investigation of i-motif stability, folding intermediates, and potential as in-situ pH sensor.Front Mol Biosci. 2022 Aug 22;9:977113. doi: 10.3389/fmolb.2022.977113. eCollection 2022. Front Mol Biosci. 2022. PMID: 36072435 Free PMC article.
-
iMab antibody binds single-stranded cytosine-rich sequences and unfolds DNA i-motifs.Nucleic Acids Res. 2024 Aug 12;52(14):8052-8062. doi: 10.1093/nar/gkae531. Nucleic Acids Res. 2024. PMID: 38908025 Free PMC article.
-
Engineering DNA quadruplexes in DNA nanostructures for biosensor construction.Nano Res. 2022;15(4):3504-3513. doi: 10.1007/s12274-021-3869-y. Epub 2021 Dec 4. Nano Res. 2022. PMID: 35401944 Free PMC article. Review.
-
Effects of Polyamine Binding on the Stability of DNA i-Motif Structures.ACS Omega. 2019 May 22;4(5):8967-8973. doi: 10.1021/acsomega.9b00784. eCollection 2019 May 31. ACS Omega. 2019. PMID: 31459985 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

