HOS1 regulates Argonaute1 by promoting transcription of the microRNA gene MIR168b in Arabidopsis
- PMID: 25619693
- PMCID: PMC4355216
- DOI: 10.1111/tpj.12772
HOS1 regulates Argonaute1 by promoting transcription of the microRNA gene MIR168b in Arabidopsis
Abstract
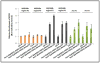
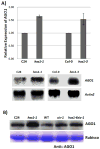
Proper accumulation and function of miRNAs is essential for plant growth and development. While core components of the miRNA biogenesis pathway and miRNA-induced silencing complex have been well characterized, cellular regulators of miRNAs remain to be fully explored. Here we report that High Expression Of Osmotically Responsive Genes1 (HOS1) is a regulator of an important miRNA, mi168a/b, that targets the Argonaute1 (AGO1) gene in Arabidopsis. HOS1 functions as an ubiquitin E3 ligase to regulate plant cold-stress responses, associates with the nuclear pores to regulate mRNA export, and regulates the circadian clock and flowering time by binding to chromatin of the flowering regulator gene Flowering Locus C (FLC). In a genetic screen for enhancers of sic-1, we isolated a loss-of-function Arabidopsis mutant of HOS1 that is defective in miRNA biogenesis. Like other hos1 mutant alleles, the hos1-7 mutant flowered early and was smaller in stature than the wild-type. Dysfunction in HOS1 reduced the abundance of miR168a/b but not of other miRNAs. In hos1 mutants, pri-MIR168b and pre-MIR168b levels were decreased, and RNA polymerase II occupancy was reduced at the promoter of MIR168b but not that of MIR168a. Chromatin immunoprecipitation assays revealed that HOS1 protein is enriched at the chromatin of the MIR168b promoter. The reduced miR168a/b level in hos1 mutants results in an increase in the mRNA and protein levels of its target gene, AGO1. Our results reveal that HOS1 regulates miR168a/b and AGO1 levels in Arabidopsis by maintaining proper transcription of MIR168b.
Keywords: Arabidopsis thaliana; HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1; MIR168b; gene expression; microRNA; transcriptional regulation.
© 2015 The Authors The Plant Journal © 2015 John Wiley & Sons Ltd.
Conflict of interest statement
The authors wish to state that there is no conflict of interest associated with any part of this manuscript.
Figures



Similar articles
-
The E3 ubiquitin ligase HOS1 regulates low ambient temperature-responsive flowering in Arabidopsis thaliana.Plant Cell Physiol. 2012 Oct;53(10):1802-14. doi: 10.1093/pcp/pcs123. Epub 2012 Sep 7. Plant Cell Physiol. 2012. PMID: 22960247
-
HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis.Plant Cell. 2013 Nov;25(11):4391-404. doi: 10.1105/tpc.113.114959. Epub 2013 Nov 19. Plant Cell. 2013. PMID: 24254125 Free PMC article.
-
The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis.Plant Cell. 2013 Nov;25(11):4378-90. doi: 10.1105/tpc.113.118364. Epub 2013 Nov 12. Plant Cell. 2013. PMID: 24220632 Free PMC article.
-
Exploring the pleiotropy of hos1.J Exp Bot. 2015 Mar;66(6):1661-71. doi: 10.1093/jxb/erv022. Epub 2015 Feb 19. J Exp Bot. 2015. PMID: 25697795 Review.
-
Beyond ubiquitination: proteolytic and nonproteolytic roles of HOS1.Trends Plant Sci. 2014 Aug;19(8):538-45. doi: 10.1016/j.tplants.2014.03.012. Epub 2014 Apr 22. Trends Plant Sci. 2014. PMID: 24768209 Review.
Cited by
-
Absence of SICKLE triggers programed cell death by disturbing alternative splicing and decay of mRNAs.Plant Physiol. 2023 Jul 3;192(3):2523-2536. doi: 10.1093/plphys/kiad192. Plant Physiol. 2023. PMID: 36974901 Free PMC article.
-
Molecular Mechanisms Underlying Freezing Tolerance in Plants: Implications for Cryopreservation.Int J Mol Sci. 2024 Sep 20;25(18):10110. doi: 10.3390/ijms251810110. Int J Mol Sci. 2024. PMID: 39337593 Free PMC article. Review.
-
Active 5' splice sites regulate the biogenesis efficiency of Arabidopsis microRNAs derived from intron-containing genes.Nucleic Acids Res. 2017 Mar 17;45(5):2757-2775. doi: 10.1093/nar/gkw895. Nucleic Acids Res. 2017. PMID: 27907902 Free PMC article.
-
CRISPR/Cas9-Mediated Knockout of HOS1 Reveals Its Role in the Regulation of Secondary Metabolism in Arabidopsis thaliana.Plants (Basel). 2021 Jan 6;10(1):104. doi: 10.3390/plants10010104. Plants (Basel). 2021. PMID: 33419060 Free PMC article.
-
An Arabidopsis Nucleoporin NUP85 modulates plant responses to ABA and salt stress.PLoS Genet. 2017 Dec 12;13(12):e1007124. doi: 10.1371/journal.pgen.1007124. eCollection 2017 Dec. PLoS Genet. 2017. PMID: 29232718 Free PMC article.
References
-
- Carmell MA, Xuan Z, Zhang M, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

