A rotary motor drives Flavobacterium gliding
- PMID: 25619763
- PMCID: PMC4319542
- DOI: 10.1016/j.cub.2014.11.045
A rotary motor drives Flavobacterium gliding
Abstract
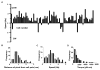
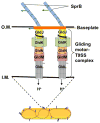
Cells of Flavobacterium johnsoniae, a rod-shaped bacterium devoid of pili or flagella, glide over glass at speeds of 2-4 μm/s [1]. Gliding is powered by a protonmotive force [2], but the machinery required for this motion is not known. Usually, cells move along straight paths, but sometimes they exhibit a reciprocal motion, attach near one pole and flip end over end, or rotate. This behavior is similar to that of a Cytophaga species described earlier [3]. Development of genetic tools for F. johnsoniae led to discovery of proteins involved in gliding [4]. These include the surface adhesin SprB that forms filaments about 160 nm long by 6 nm in diameter, which, when labeled with a fluorescent antibody [2] or a latex bead [5], are seen to move longitudinally down the length of a cell, occasionally shifting positions to the right or the left. Evidently, interaction of these filaments with a surface produces gliding. To learn more about the gliding motor, we sheared cells to reduce the number and size of SprB filaments and tethered cells to glass by adding anti-SprB antibody. Cells spun about fixed points, mostly counterclockwise, rotating at speeds of 1 Hz or more. The torques required to sustain such speeds were large, comparable to those generated by the flagellar rotary motor. However, we found that a gliding motor runs at constant speed rather than at constant torque. Now, there are three rotary motors powered by protonmotive force: the bacterial flagellar motor, the Fo ATP synthase, and the gliding motor.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures

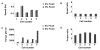
References
-
- Jarrell KF, McBride MJ. The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol. 2008;6:466–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

