Transcriptomic analysis reveals the roles of microtubule-related genes and transcription factors in fruit length regulation in cucumber (Cucumis sativus L.)
- PMID: 25619948
- PMCID: PMC5379036
- DOI: 10.1038/srep08031
Transcriptomic analysis reveals the roles of microtubule-related genes and transcription factors in fruit length regulation in cucumber (Cucumis sativus L.)
Abstract
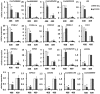
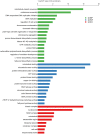
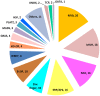
Cucumber (Cucumis sativus L.) fruit is a type of fleshy fruit that is harvested immaturely. Early fruit development directly determines the final fruit length and diameter, and consequently the fruit yield and quality. Different cucumber varieties display huge variations of fruit length, but how fruit length is determined at the molecular level remains poorly understood. To understand the genes and gene networks that regulate fruit length in cucumber, high throughout RNA-Seq data were used to compare the transcriptomes of early fruit from two near isogenic lines with different fruit lengths. 3955 genes were found to be differentially expressed, among which 2368 genes were significantly up-regulated and 1587 down-regulated in the line with long fruit. Microtubule and cell cycle related genes were dramatically activated in the long fruit, and transcription factors were implicated in the fruit length regulation in cucumber. Thus, our results built a foundation for dissecting the molecular mechanism of fruit length control in cucumber, a key agricultural trait of significant economic importance.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

