Local error estimates dramatically improve the utility of homology models for solving crystal structures by molecular replacement
- PMID: 25619999
- PMCID: PMC4321884
- DOI: 10.1016/j.str.2014.11.020
Local error estimates dramatically improve the utility of homology models for solving crystal structures by molecular replacement
Abstract
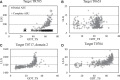
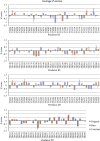
Predicted structures submitted for CASP10 have been evaluated as molecular replacement models against the corresponding sets of structure factor amplitudes. It has been found that the log-likelihood gain score computed for each prediction correlates well with common structure quality indicators but is more sensitive when the accuracy of the models is high. In addition, it was observed that using coordinate error estimates submitted by predictors to weight the model can improve its utility in molecular replacement dramatically, and several groups have been identified who reliably provide accurate error estimates that could be used to extend the application of molecular replacement for low-homology cases.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Berman H., Henrick K., Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. - PubMed
-
- Boniecki M., Rotkiewicz P., Skolnick J., Kolinski A. Protein fragment reconstruction using various modeling techniques. J. Comput. Aided Mol. Des. 2003;17:725–738. - PubMed
-
- Buenavista M.T., Roche D.B., McGuffin L.J. Improvement of 3D protein models using multiple templates guided by single-template model quality assessment. Bioinformatics. 2012;28:1851–1857. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

