Lupus risk variants in the PXK locus alter B-cell receptor internalization
- PMID: 25620976
- PMCID: PMC4288052
- DOI: 10.3389/fgene.2014.00450
Lupus risk variants in the PXK locus alter B-cell receptor internalization
Abstract
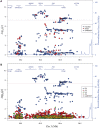
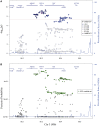
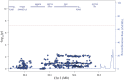
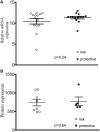
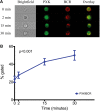
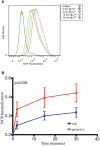
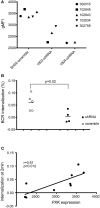
Genome wide association studies have identified variants in PXK that confer risk for humoral autoimmune diseases, including systemic lupus erythematosus (SLE or lupus), rheumatoid arthritis and more recently systemic sclerosis. While PXK is involved in trafficking of epidermal growth factor Receptor (EGFR) in COS-7 cells, mechanisms linking PXK to lupus pathophysiology have remained undefined. In an effort to uncover the mechanism at this locus that increases lupus-risk, we undertook a fine-mapping analysis in a large multi-ancestral study of lupus patients and controls. We define a large (257kb) common haplotype marking a single causal variant that confers lupus risk detected only in European ancestral populations and spans the promoter through the 3' UTR of PXK. The strongest association was found at rs6445972 with P < 4.62 × 10(-10), OR 0.81 (0.75-0.86). Using stepwise logistic regression analysis, we demonstrate that one signal drives the genetic association in the region. Bayesian analysis confirms our results, identifying a 95% credible set consisting of 172 variants spanning 202 kb. Functionally, we found that PXK operates on the B-cell antigen receptor (BCR); we confirmed that PXK influenced the rate of BCR internalization. Furthermore, we demonstrate that individuals carrying the risk haplotype exhibited a decreased rate of BCR internalization, a process known to impact B cell survival and cell fate. Taken together, these data define a new candidate mechanism for the genetic association of variants around PXK with lupus risk and highlight the regulation of intracellular trafficking as a genetically regulated pathway mediating human autoimmunity.
Keywords: B cells; BCR; PXK; fine-mapping; lupus.
Figures
Similar articles
-
PXK locus in systemic lupus erythematosus: fine mapping and functional analysis reveals novel susceptibility gene ABHD6.Ann Rheum Dis. 2015 Mar;74(3):e14. doi: 10.1136/annrheumdis-2013-204909. Epub 2014 Feb 17. Ann Rheum Dis. 2015. PMID: 24534757
-
Population differences in SLE susceptibility genes: STAT4 and BLK, but not PXK, are associated with systemic lupus erythematosus in Hong Kong Chinese.Genes Immun. 2009 Apr;10(3):219-26. doi: 10.1038/gene.2009.1. Epub 2009 Feb 19. Genes Immun. 2009. PMID: 19225526
-
Replication of GWAS-identified systemic lupus erythematosus susceptibility genes affirms B-cell receptor pathway signalling and strengthens the role of IRF5 in disease susceptibility in a Northern European population.Rheumatology (Oxford). 2012 Jan;51(1):87-92. doi: 10.1093/rheumatology/ker263. Epub 2011 Oct 27. Rheumatology (Oxford). 2012. PMID: 22039224 Review.
-
Mechanistic Characterization of RASGRP1 Variants Identifies an hnRNP-K-Regulated Transcriptional Enhancer Contributing to SLE Susceptibility.Front Immunol. 2019 May 20;10:1066. doi: 10.3389/fimmu.2019.01066. eCollection 2019. Front Immunol. 2019. PMID: 31164884 Free PMC article.
-
What can we learn from genetic studies of systemic lupus erythematosus? Implications of genetic heterogeneity among populations in SLE.Lupus. 2010 Oct;19(12):1452-9. doi: 10.1177/0961203310370350. Lupus. 2010. PMID: 20947557 Review.
Cited by
-
A plausibly causal functional lupus-associated risk variant in the STAT1-STAT4 locus.Hum Mol Genet. 2018 Jul 1;27(13):2392-2404. doi: 10.1093/hmg/ddy140. Hum Mol Genet. 2018. PMID: 29912393 Free PMC article. Clinical Trial.
-
CASCADE: high-throughput characterization of regulatory complex binding altered by non-coding variants.Cell Genom. 2022 Feb 9;2(2):100098. doi: 10.1016/j.xgen.2022.100098. Cell Genom. 2022. PMID: 35252945 Free PMC article.
-
Lymphocytes Change Their Phenotype and Function in Systemic Lupus Erythematosus and Lupus Nephritis.Int J Mol Sci. 2024 Oct 10;25(20):10905. doi: 10.3390/ijms252010905. Int J Mol Sci. 2024. PMID: 39456692 Free PMC article. Review.
-
Machine learning approaches to predict lupus disease activity from gene expression data.Sci Rep. 2019 Jul 3;9(1):9617. doi: 10.1038/s41598-019-45989-0. Sci Rep. 2019. PMID: 31270349 Free PMC article.
-
The Role of Genetic Risk Factors in Pathogenesis of Childhood-Onset Systemic Lupus Erythematosus.Curr Issues Mol Biol. 2023 Jul 17;45(7):5981-6002. doi: 10.3390/cimb45070378. Curr Issues Mol Biol. 2023. PMID: 37504294 Free PMC article. Review.
References
-
- Alhouayek M., Masquelier J., Cani P. D., Lambert D. M., Muccioli G. G. (2013). Implication of the anti-inflammatory bioactive lipid prostaglandin D2-glycerol ester in the control of macrophage activation and inflammation by ABHD6. Proc. Natl. Acad. Sci. U.S.A. 110, 17558–17563. 10.1073/pnas.1314017110 - DOI - PMC - PubMed
Grants and funding
- P60 AR053308/AR/NIAMS NIH HHS/United States
- U54 GM104938/GM/NIGMS NIH HHS/United States
- R21 AR065626/AR/NIAMS NIH HHS/United States
- UL1 TR000154/TR/NCATS NIH HHS/United States
- P30 DK078392/DK/NIDDK NIH HHS/United States
- T32 GM063483/GM/NIGMS NIH HHS/United States
- P30 CA046934/CA/NCI NIH HHS/United States
- R01 AR043814/AR/NIAMS NIH HHS/United States
- R37 AI024717/AI/NIAID NIH HHS/United States
- UL1 RR029882/RR/NCRR NIH HHS/United States
- R01 AR043727/AR/NIAMS NIH HHS/United States
- UL1 TR001425/TR/NCATS NIH HHS/United States
- I01 BX001834/BX/BLRD VA/United States
- UL1 TR001082/TR/NCATS NIH HHS/United States
- K24 AI078004/AI/NIAID NIH HHS/United States
- P30 DK090971/DK/NIDDK NIH HHS/United States
- R21 AI070304/AI/NIAID NIH HHS/United States
- P01 AI083194/AI/NIAID NIH HHS/United States
- P01 AR049084/AR/NIAMS NIH HHS/United States
- P30 AR047363/AR/NIAMS NIH HHS/United States
- P60 AR062755/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

