Whole-body transcriptome of selectively bred, resistant-, control-, and susceptible-line rainbow trout following experimental challenge with Flavobacterium psychrophilum
- PMID: 25620978
- PMCID: PMC4288049
- DOI: 10.3389/fgene.2014.00453
Whole-body transcriptome of selectively bred, resistant-, control-, and susceptible-line rainbow trout following experimental challenge with Flavobacterium psychrophilum
Abstract
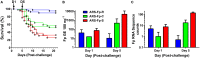
Genetic improvement for enhanced disease resistance in fish is an increasingly utilized approach to mitigate endemic infectious disease in aquaculture. In domesticated salmonid populations, large phenotypic variation in disease resistance has been identified but the genetic basis for altered responsiveness remains unclear. We previously reported three generations of selection and phenotypic validation of a bacterial cold water disease (BCWD) resistant line of rainbow trout, designated ARS-Fp-R. This line has higher survival after infection by either standardized laboratory challenge or natural challenge as compared to two reference lines, designated ARS-Fp-C (control) and ARS-Fp-S (susceptible). In this study, we utilized 1.1 g fry from the three genetic lines and performed RNA-seq to measure transcript abundance from the whole body of naive and Flavobacterium psychrophilum infected fish at day 1 (early time-point) and at day 5 post-challenge (onset of mortality). Sequences from 24 libraries were mapped onto the rainbow trout genome reference transcriptome of 46,585 predicted protein coding mRNAs that included 2633 putative immune-relevant gene transcripts. A total of 1884 genes (4.0% genome) exhibited differential transcript abundance between infected and mock-challenged fish (FDR < 0.05) that included chemokines, complement components, tnf receptor superfamily members, interleukins, nod-like receptor family members, and genes involved in metabolism and wound healing. The largest number of differentially expressed genes occurred on day 5 post-infection between naive and challenged ARS-Fp-S line fish correlating with high bacterial load. After excluding the effect of infection, we identified 21 differentially expressed genes between the three genetic lines. In summary, these data indicate global transcriptome differences between genetic lines of naive animals as well as differentially regulated transcriptional responses to infection.
Keywords: Flavobacterium psychrophilum; aquaculture; bacterial cold water disease; disease resistance; immune gene; rainbow trout genome; selective breeding; tnfrsf.
Figures






References
-
- Barnes M. E., Brown M. L. (2011). A review of Flavobacterium psychrophilum biology, clinical signs, and bacterial cold water disease prevention and treatment. Open Fish Sci. J. 4, 40–48 10.2174/1874401X01104010040 - DOI
-
- Bayne C. J., Gerwick L., Fujiki K., Nakao M., Yano T. (2001). Immune-relevant (including acute phase) genes identified in the livers of rainbow trout, Oncorhynchus mykiss, by means of suppression subtractive hybridization. Dev. Comp. Immunol. 25, 205–217. 10.1016/S0145-305X(00)00057-4 - DOI - PubMed
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical approach to multiple testing. J. R. Stat. Soc. B 57, 289–300.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

