Large multiallelic copy number variations in humans
- PMID: 25621458
- PMCID: PMC4405206
- DOI: 10.1038/ng.3200
Large multiallelic copy number variations in humans
Abstract
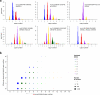
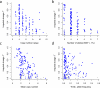
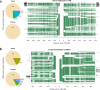
Thousands of genomic segments appear to be present in widely varying copy numbers in different human genomes. We developed ways to use increasingly abundant whole-genome sequence data to identify the copy numbers, alleles and haplotypes present at most large multiallelic CNVs (mCNVs). We analyzed 849 genomes sequenced by the 1000 Genomes Project to identify most large (>5-kb) mCNVs, including 3,878 duplications, of which 1,356 appear to have 3 or more segregating alleles. We find that mCNVs give rise to most human variation in gene dosage-seven times the combined contribution of deletions and biallelic duplications-and that this variation in gene dosage generates abundant variation in gene expression. We describe 'runaway duplication haplotypes' in which genes, including HPR and ORM1, have mutated to high copy number on specific haplotypes. We also describe partially successful initial strategies for analyzing mCNVs via imputation and provide an initial data resource to support such analyses.
Figures



References
-
- Weiss LA, et al. Association between microdeletion and microduplication at 16p11.2 and autism. N Engl J Med. 2008;358:667–75. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

