Convergent evolution of the genomes of marine mammals
- PMID: 25621460
- PMCID: PMC4644735
- DOI: 10.1038/ng.3198
Convergent evolution of the genomes of marine mammals
Abstract
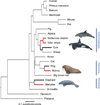
Marine mammals from different mammalian orders share several phenotypic traits adapted to the aquatic environment and therefore represent a classic example of convergent evolution. To investigate convergent evolution at the genomic level, we sequenced and performed de novo assembly of the genomes of three species of marine mammals (the killer whale, walrus and manatee) from three mammalian orders that share independently evolved phenotypic adaptations to a marine existence. Our comparative genomic analyses found that convergent amino acid substitutions were widespread throughout the genome and that a subset of these substitutions were in genes evolving under positive selection and putatively associated with a marine phenotype. However, we found higher levels of convergent amino acid substitutions in a control set of terrestrial sister taxa to the marine mammals. Our results suggest that, whereas convergent molecular evolution is relatively common, adaptive molecular convergence linked to phenotypic convergence is comparatively rare.
Figures

References
-
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–114. - PubMed
-
- Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335:457–461. - PubMed
-
- Stern DL. The genetic causes of convergent evolution. Nature Rev. Genet. 2013;14:751–764. - PubMed
-
- Stewart CB, Schilling JW, Wilson AC. Adaptive evolution in the stomach lysozymes of foregut fermenters. Nature. 1987;330:401–404. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

