Improving target cell specificity using a novel monovalent bispecific IgG design
- PMID: 25621507
- PMCID: PMC4622537
- DOI: 10.1080/19420862.2015.1007816
Improving target cell specificity using a novel monovalent bispecific IgG design
Erratum in
-
Corrigendum.MAbs. 2016 May-Jun;8(4):839. doi: 10.1080/19420862.2016.1175161. MAbs. 2016. PMID: 27142173 Free PMC article. No abstract available.
Abstract
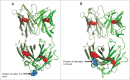
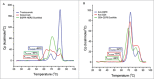
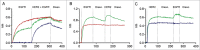
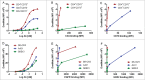
Monovalent bispecific IgGs cater to a distinct set of mechanisms of action but are difficult to engineer and manufacture because of complexities associated with correct heavy and light chain pairing. We have created a novel design, "DuetMab," for efficient production of these molecules. The platform uses knobs-into-holes (KIH) technology for heterodimerization of 2 distinct heavy chains and increases the efficiency of cognate heavy and light chain pairing by replacing the native disulfide bond in one of the CH1-CL interfaces with an engineered disulfide bond. Using two pairs of antibodies, cetuximab (anti-EGFR) and trastuzumab (anti-HER2), and anti-CD40 and anti-CD70 antibodies, we demonstrate that DuetMab antibodies can be produced in a highly purified and active form, and show for the first time that monovalent bispecific IgGs can concurrently bind both antigens on the same cell. This last property compensates for the loss of avidity brought about by monovalency and improves selectivity toward the target cell.
Keywords: ADCC, antibody-dependent cell-mediated cytotoxicity; Biotechnology; CDR, complementarity determining region; CH1, 2 and 3-heavy chain constant domain 1, 2 and 3; CL-, light chain constant domain; DSC-differential scanning calorimetry; E:T, ratio of effector to target cells; EGFR; EGFR, epidermal growth factor receptor; FcRn, neonatal Fc receptor; FcγR, receptor for IgG Fc; HER2; IGFR, insulin like growth factor receptor; IL-6, interleukin 6; IgG, Immunoglobulin G; PNGase, protein N-glycanase; Q1q, first component of complement 1; RAGE, receptor for advanced glycosylation; antibody engineering; bispecific antibody; cancer; disulfide; mAbs, monoclonal antibodies; multi-targeting.
Figures



References
-
- Kontermann RE. Alternative antibody formats. Curr Opin Mol Ther 2010; 12:176-83; PMID:20373261 - PubMed
-
- Kontermann R. Dual targeting strategies with bispecific antibodies. MAbs 2012; 4:182-97; PMID:22453100; http://dx.doi.org/10.4161/mabs.4.2.19000 - DOI - PMC - PubMed
-
- Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol 2010; 10:301-16; PMID:20414204; http://dx.doi.org/10.1038/nri2761 - DOI - PubMed
-
- Marvin JS, Zhu Z. Recombinant approaches to IgG-like bispecific antibodies. Acta Pharmacol Sin 2005; 26:649-58; PMID:15916729; http://dx.doi.org/10.1111/j.1745-7254.2005.00119.x - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
