Interactions of amino acid side-chain analogs within membrane environments
- PMID: 25621811
- PMCID: PMC4420737
- DOI: 10.1021/jp511712u
Interactions of amino acid side-chain analogs within membrane environments
Abstract
The interactions among four amino acid analog pairs (Asn, Ser, Phe, and Val) within the membrane environment were investigated using umbrella sampling molecular dynamics simulations. The results confirm generally expected qualitative trends of preferential association of polar compounds inside the membrane vs preferential interaction of hydrophobic compounds outside the membrane. Furthermore, correlations between amino acid interactions, membrane insertion, and membrane deformations are discussed and a detailed analysis of pair interaction energies is presented. A comparison of the energetics obtained from explicit lipid simulations with those from implicit membrane models reveals significant deviations and an improved parametrization of the heterogeneous dielectric generalized Born implicit model is provided that partially corrects for deficiencies in the implicit membrane model when compared with the new reference data from this study.
Figures

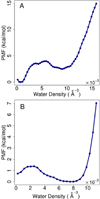

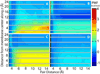
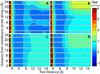
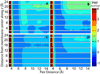


References
-
- Bennett WFD, Tieleman DP. Computer simulations of lipid membrane domains. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2013;1828(8):1765–1776. - PubMed
-
- Andersen OS, Koeppe RE. Bilayer Thickness and Membrane Protein Function: An Energetic Perspective. Annual Review of Biophysics and Biomolecular Structure. 2007;36(1):107–130. - PubMed
-
- Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111(27):7812–7824. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

