The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4
- PMID: 25621826
- PMCID: PMC4333215
- DOI: 10.1038/ni.3093
The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4
Abstract
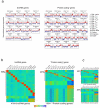
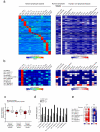
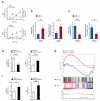
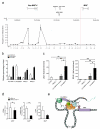
Long noncoding RNAs are emerging as important regulators of cellular functions, but little is known of their role in the human immune system. Here we investigated long intergenic noncoding RNAs (lincRNAs) in 13 subsets of T lymphocytes and B lymphocytes by next-generation sequencing-based RNA sequencing (RNA-seq analysis) and de novo transcriptome reconstruction. We identified over 500 previously unknown lincRNAs and described lincRNA signatures. Expression of linc-MAF-4, a chromatin-associated lincRNA specific to the TH1 subset of helper T cells, was inversely correlated with expression of MAF, a TH2-associated transcription factor. Downregulation of linc-MAF-4 skewed T cell differentiation toward the TH2 phenotype. We identified a long-distance interaction between the genomic regions of the gene encoding linc-MAF-4 and MAF, where linc-MAF-4 associated with the chromatin modifiers LSD1 and EZH2; this suggested that linc-MAF-4 regulated MAF transcription through the recruitment of chromatin modifiers. Our results demonstrate a key role for lincRNA in T lymphocyte differentiation.
Figures


Comment in
-
LincRNA signatures in human lymphocytes.Nat Immunol. 2015 Mar;16(3):220-2. doi: 10.1038/ni.3106. Nat Immunol. 2015. PMID: 25689433 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

