Conversations between kingdoms: small RNAs
- PMID: 25622136
- PMCID: PMC4387066
- DOI: 10.1016/j.copbio.2014.12.025
Conversations between kingdoms: small RNAs
Abstract
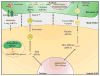
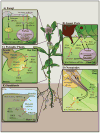
Humans, animals, and plants are constantly under attack from pathogens and pests, resulting in severe consequences on global human health and crop production. Small RNA (sRNA)-mediated RNA interference (RNAi) is a conserved regulatory mechanism that is involved in almost all eukaryotic cellular processes, including host immunity and pathogen virulence. Recent evidence supports the significant contribution of sRNAs and RNAi to the communication between hosts and some eukaryotic pathogens, pests, parasites, or symbiotic microorganisms. Mobile silencing signals—most likely sRNAs—are capable of translocating from the host to its interacting organism, and vice versa. In this review, we will provide an overview of sRNA communications between different kingdoms, with a primary focus on the advances in plant-pathogen interaction systems.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
References
-
- Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013;197:394–404. - PubMed
-
- Yang L, Huang H. Roles of small RNAs in plant disease resistance. J Integr Plant Biol. 2014;56:962–970. - PubMed
-
- Weiberg A, Wang M, Bellinger M, Jin H. Small RNAs: A New Paradigm in Plant-Microbe Interactions. Annu Rev Phytopathol. 2014;52:495–516. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

