Expression of the SIRT2 gene and its relationship with body size traits in Qinchuan cattle (Bos taurus)
- PMID: 25622258
- PMCID: PMC4346846
- DOI: 10.3390/ijms16022458
Expression of the SIRT2 gene and its relationship with body size traits in Qinchuan cattle (Bos taurus)
Abstract
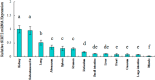
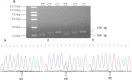
Silent information regulator 2 (SIRT2) is a member of the sirtuin family of class III NAD (nicotinamide adenine dinucleotide)-dependent protein deacetylases and may regulate senescence, metabolism and apoptosis. The aims of this study were to investigate whether the SIRT2 gene could be used as a candidate gene in the breeding of Qinchuan cattle. Real-time polymerase chain reaction (RT-PCR) results showed that among all types of tissue that were analyzed, the highest mRNA expression levels of the gene were found in subcutaneous fat. DNA sequencing of 468 individual Qinchuan cattle identified two novel, single nucleotide polymorphisms (g.19501 C > T and g.19518 C > T) in the 3' untranslated region (3'UTR) of the SIRT2 gene. The frequencies of SNP g.19501 C > T and g.19518 C > T were in Hardy-Weinberg disequilibrium in all the samples (chi-square test, χ2 < χ0.052). An association analysis showed that the two loci were significantly correlated with some body size traits and the H2H2 (-CT-CT-) diplotypes performed better than other combinations. These results indicated that the variations in the SIRT2 gene and their corresponding genotypes may be considered as molecular markers for economic traits in cattle breeding.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

