Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation
- PMID: 25622295
- PMCID: PMC4351163
- DOI: 10.1038/nsmb.2957
Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation
Abstract
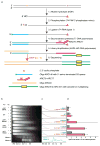
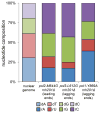
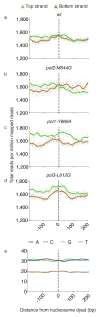
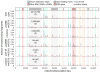
Ribonucleotides are frequently incorporated into DNA during replication in eukaryotes. Here we map genome-wide distribution of these ribonucleotides as markers of replication enzymology in budding yeast, using a new 5' DNA end-mapping method, hydrolytic end sequencing (HydEn-seq). HydEn-seq of DNA from ribonucleotide excision repair-deficient strains reveals replicase- and strand-specific patterns of ribonucleotides in the nuclear genome. These patterns support the roles of DNA polymerases α and δ in lagging-strand replication and of DNA polymerase ɛ in leading-strand replication. They identify replication origins, termination zones and variations in ribonucleotide incorporation frequency across the genome that exceed three orders of magnitude. HydEn-seq also reveals strand-specific 5' DNA ends at mitochondrial replication origins, thus suggesting unidirectional replication of a circular genome. Given the conservation of enzymes that incorporate and process ribonucleotides in DNA, HydEn-seq can be used to track replication enzymology in other organisms.
Conflict of interest statement
The authors declare no competing financial interests. The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

