Transcriptome analysis of the hormone-sensing cells in mammary epithelial reveals dynamic changes in early pregnancy
- PMID: 25623114
- PMCID: PMC4314744
- DOI: 10.1186/s12861-015-0058-9
Transcriptome analysis of the hormone-sensing cells in mammary epithelial reveals dynamic changes in early pregnancy
Abstract
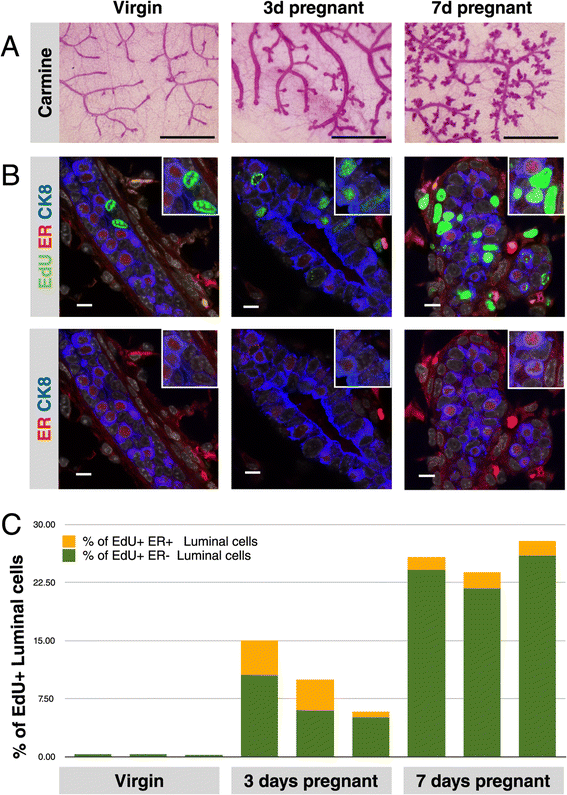
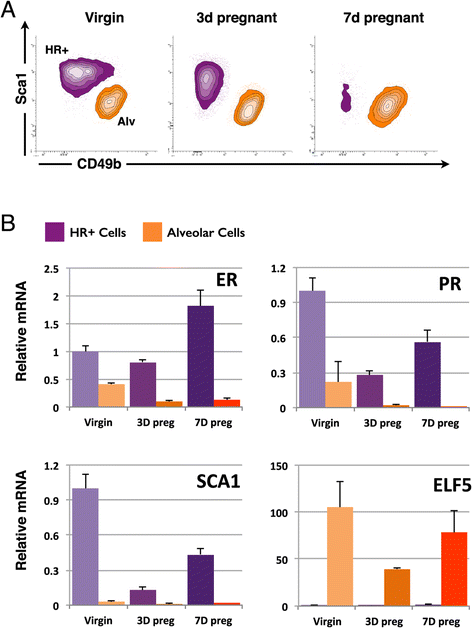
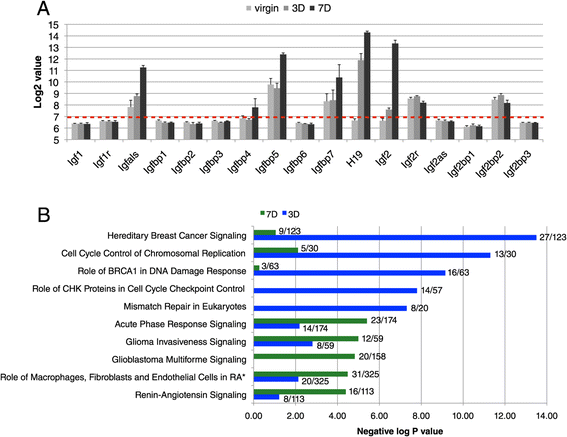
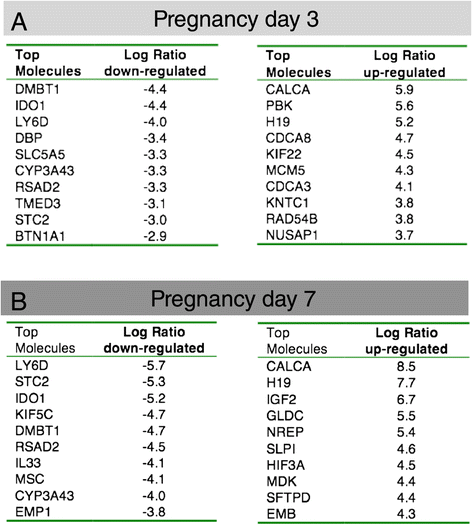
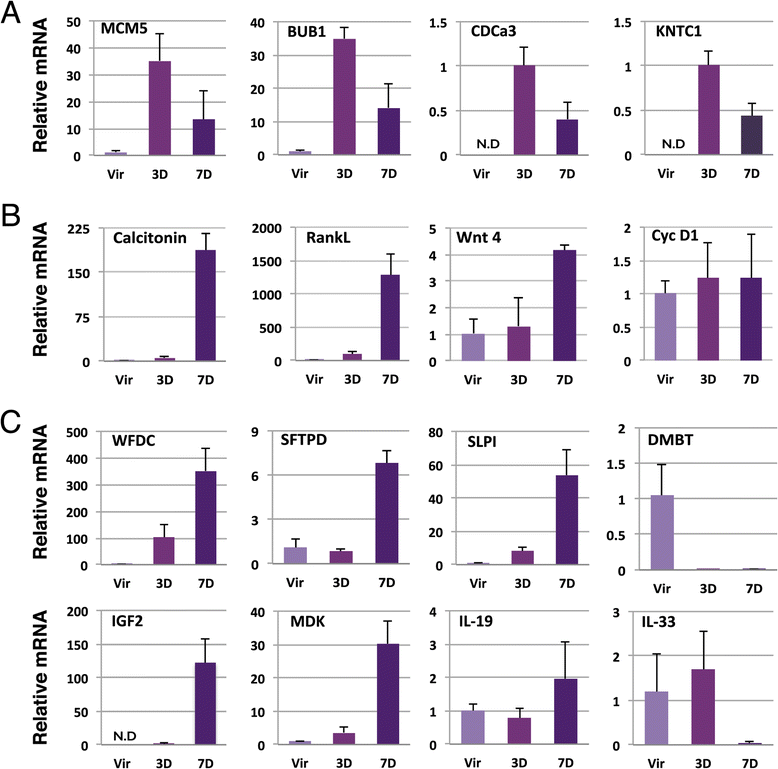
Background: Alveoli, the milk-producing units of the mammary gland, are generated during pregnancy by collaboration of different epithelial cell types. We present the first analysis of transcriptional changes within the hormone sensing population during pregnancy. Hormone-receptor positive (HR+) cells play a key role in the initiation of alveologenesis as they sense systemic hormonal changes and translate these into local instructions for neighboring HR- cells. We recently showed that IGF2 is produced specifically by HR+ cells in early pregnancy, but is undetectable in the virgin state. Here, we define the transcriptome of HR+ cells in early pregnancy with the aim to elucidate additional changes that are unique for this dynamic developmental time window.
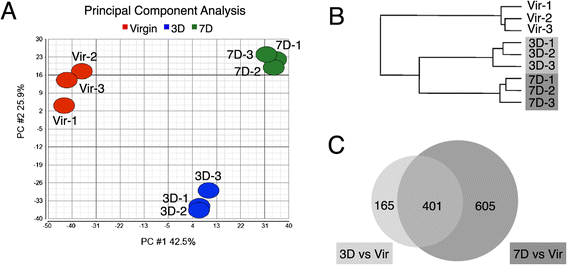
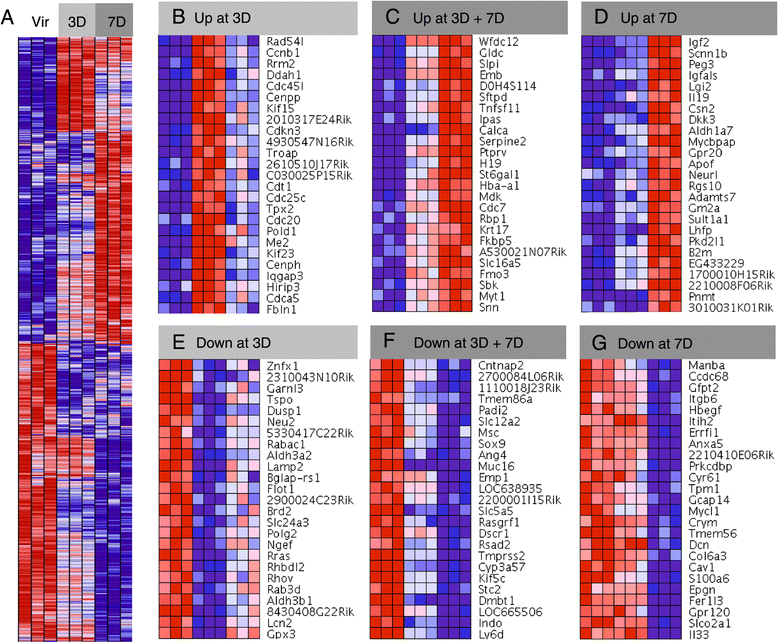
Results: We harvested mammary glands from virgin, 3-day and 7-day pregnant mice and isolated a few hundred hormone-sensing cells per animal by FACS for microarray analysis. There was a high concordance between animals with a clear induction of cell cycle progression genes at day 3 of pregnancy and molecules involved in paracrine signalling at day 7.
Conclusions: These findings underscore the proliferative capacity of HR+ cells upon specific stimuli and elucidate developmentally-restricted changes in cellular communication. Since the majority of breast cancers are HR+, with a variable proportion of HR+ cells per tumor, we anticipate that this data set will aid further studies into the regulation of HR+ cell proliferation and the role of heterotypic signalling within tumors.
Figures
Similar articles
-
Sustained trophism of the mammary gland is sufficient to accelerate and synchronize development of ErbB2/Neu-induced tumors.Oncogene. 2006 Jun 1;25(23):3325-34. doi: 10.1038/sj.onc.1209365. Epub 2006 Jan 23. Oncogene. 2006. PMID: 16434967 Free PMC article.
-
Functional development of the adult ovine mammary gland--insights from gene expression profiling.BMC Genomics. 2015 Oct 5;16:748. doi: 10.1186/s12864-015-1947-9. BMC Genomics. 2015. PMID: 26437771 Free PMC article.
-
Selective segregation of DNA strands persists in long-label-retaining mammary cells during pregnancy.Breast Cancer Res. 2008;10(5):R90. doi: 10.1186/bcr2188. Epub 2008 Oct 24. Breast Cancer Res. 2008. PMID: 18950502 Free PMC article.
-
How pregnancy at early age protects against breast cancer.Trends Mol Med. 2014 Mar;20(3):143-53. doi: 10.1016/j.molmed.2013.11.002. Epub 2013 Dec 16. Trends Mol Med. 2014. PMID: 24355762 Review.
-
Hormonal approach to breast cancer prevention.J Cell Biochem Suppl. 2000;34:1-6. J Cell Biochem Suppl. 2000. PMID: 10762007 Review.
Cited by
-
The Use of "Omics" in Lactation Research in Dairy Cows.Int J Mol Sci. 2017 May 5;18(5):983. doi: 10.3390/ijms18050983. Int J Mol Sci. 2017. PMID: 28475129 Free PMC article. Review.
-
Chi-miR-3031 regulates beta-casein via the PI3K/AKT-mTOR signaling pathway in goat mammary epithelial cells (GMECs).BMC Vet Res. 2018 Nov 27;14(1):369. doi: 10.1186/s12917-018-1695-6. BMC Vet Res. 2018. PMID: 30482199 Free PMC article.
-
Hormone-sensing mammary epithelial progenitors: emerging identity and hormonal regulation.J Mammary Gland Biol Neoplasia. 2015 Jun;20(1-2):75-91. doi: 10.1007/s10911-015-9344-1. Epub 2015 Sep 21. J Mammary Gland Biol Neoplasia. 2015. PMID: 26390871 Review.
-
ERrrr…where are the progenitors? Hormone receptors and mammary cell heterogeneity.J Mammary Gland Biol Neoplasia. 2015 Jun;20(1-2):63-73. doi: 10.1007/s10911-015-9336-1. Epub 2015 Jul 21. J Mammary Gland Biol Neoplasia. 2015. PMID: 26193872 Free PMC article. Review.
-
Form and function: how estrogen and progesterone regulate the mammary epithelial hierarchy.J Mammary Gland Biol Neoplasia. 2015 Jun;20(1-2):9-25. doi: 10.1007/s10911-015-9337-0. Epub 2015 Jul 19. J Mammary Gland Biol Neoplasia. 2015. PMID: 26188694 Free PMC article. Review.
References
-
- Clarke RB, Howell A, Potten CS, Anderson E. Dissociation between steroid receptor expression and cell proliferation in the human breast. Cancer Res. 1997;57:4987–91. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

