Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder
- PMID: 25623948
- PMCID: PMC4414679
- DOI: 10.1038/mp.2014.176
Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder
Abstract
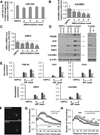
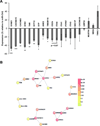
Bipolar disorder (BD) is a heritable neuropsychiatric disorder with largely unknown pathogenesis. Given their prominent role in brain function and disease, we hypothesized that microRNAs (miRNAs) might be of importance for BD. Here we show that levels of miR-34a, which is predicted to target multiple genes implicated as genetic risk factors for BD, are increased in postmortem cerebellar tissue from BD patients, as well as in BD patient-derived neuronal cultures generated by reprogramming of human fibroblasts into induced neurons or into induced pluripotent stem cells (iPSCs) subsequently differentiated into neurons. Of the predicted miR-34a targets, we validated the BD risk genes ankyrin-3 (ANK3) and voltage-dependent L-type calcium channel subunit beta-3 (CACNB3) as direct miR-34a targets. Using human iPSC-derived neuronal progenitor cells, we further show that enhancement of miR-34a expression impairs neuronal differentiation, expression of synaptic proteins and neuronal morphology, whereas reducing endogenous miR-34a expression enhances dendritic elaboration. Taken together, we propose that miR-34a serves as a critical link between multiple etiological factors for BD and its pathogenesis through the regulation of a molecular network essential for neuronal development and synaptogenesis.
Figures


References
-
- Ketter TA. Diagnostic features, prevalence, and impact of bipolar disorder. The Journal of clinical psychiatry. 2010;71(6):e14. - PubMed
-
- Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. 2013;381(9878):1654–1662. - PubMed
-
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature reviews Genetics. 2010;11(9):597–610. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

