Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues
- PMID: 25624062
- PMCID: PMC4306919
- DOI: 10.1038/srep08057
Correlation of circular RNA abundance with proliferation--exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues
Abstract
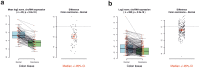
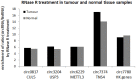
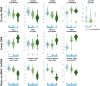
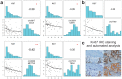
Circular RNAs are a recently (re-)discovered abundant RNA species with presumed function as miRNA sponges, thus part of the competing endogenous RNA network. We analysed the expression of circular and linear RNAs and proliferation in matched normal colon mucosa and tumour tissues. We predicted >1,800 circular RNAs and proved the existence of five randomly chosen examples using RT-qPCR. Interestingly, the ratio of circular to linear RNA isoforms was always lower in tumour compared to normal colon samples and even lower in colorectal cancer cell lines. Furthermore, this ratio correlated negatively with the proliferation index. The correlation of global circular RNA abundance (the circRNA index) and proliferation was validated in a non-cancerous proliferative disease, idiopathic pulmonary fibrosis, ovarian cancer cells compared to cultured normal ovarian epithelial cells, and 13 normal human tissues. We are the first to report a global reduction of circular RNA abundance in colorectal cancer cell lines and cancer compared to normal tissues and discovered a negative correlation of global circular RNA abundance and proliferation. This negative correlation seems to be a general principle in human tissues as validated with three different settings. Finally, we present a simple model how circular RNAs could accumulate in non-proliferating cells.
Figures


References
-
- Nigro J. M. et al. Scrambled exons. Cell 64, 607–613 (1991). - PubMed
-
- Memczak S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013). - PubMed
-
- Hansen T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

