Node-pore sensing enables label-free surface-marker profiling of single cells
- PMID: 25625182
- PMCID: PMC4350414
- DOI: 10.1021/ac504613b
Node-pore sensing enables label-free surface-marker profiling of single cells
Abstract
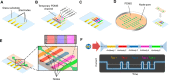
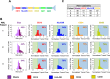
Flow cytometry is a ubiquitous, multiparametric method for characterizing cellular populations. However, this method can grow increasingly complex with the number of proteins that need to be screened simultaneously: spectral emission overlap of fluorophores and the subsequent need for compensation, lengthy sample preparation, and multiple control tests that need to be performed separately must all be considered. These factors lead to increased costs, and consequently, flow cytometry is performed in core facilities with a dedicated technician operating the instrument. Here, we describe a low-cost, label-free microfluidic method that can determine the phenotypic profiles of single cells. Our method employs Node-Pore Sensing to measure the transit times of cells as they interact with a series of different antibodies, each corresponding to a specific cell-surface antigen, that have been functionalized in a single microfluidic channel. We demonstrate the capabilities of our method not only by screening two acute promyelocytic leukemia human cells lines (NB4 and AP-1060) for myeloid antigens, CD13, CD14, CD15, and CD33, simultaneously, but also by distinguishing a mixture of cells of similar size—AP-1060 and NALM-1—based on surface markers CD13 and HLA-DR. Furthermore, we show that our method can screen complex subpopulations in clinical samples: we successfully identified the blast population in primary human bone marrow samples from patients with acute myeloid leukemia and screened these cells for CD13, CD34, and HLA-DR. We show that our label-free method is an affordable, highly sensitive, and user-friendly technology that has the potential to transform cellular screening at the benchside.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

