What RNA World? Why a Peptide/RNA Partnership Merits Renewed Experimental Attention
- PMID: 25625599
- PMCID: PMC4390853
- DOI: 10.3390/life5010294
What RNA World? Why a Peptide/RNA Partnership Merits Renewed Experimental Attention
Abstract
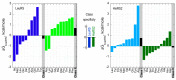
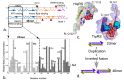
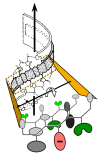
We review arguments that biology emerged from a reciprocal partnership in which small ancestral oligopeptides and oligonucleotides initially both contributed rudimentary information coding and catalytic rate accelerations, and that the superior information-bearing qualities of RNA and the superior catalytic potential of proteins emerged from such complexes only with the gradual invention of the genetic code. A coherent structural basis for that scenario was articulated nearly a decade before the demonstration of catalytic RNA. Parallel hierarchical catalytic repertoires for increasingly highly conserved sequences from the two synthetase classes now increase the likelihood that they arose as translation products from opposite strands of a single gene. Sense/antisense coding affords a new bioinformatic metric for phylogenetic relationships much more distant than can be reconstructed from multiple sequence alignments of a single superfamily. Evidence for distinct coding properties in tRNA acceptor stems and anticodons, and experimental demonstration that the two synthetase family ATP binding sites can indeed be coded by opposite strands of the same gene supplement these biochemical and bioinformatic data, establishing a solid basis for key intermediates on a path from simple, stereochemically coded, reciprocally catalytic peptide/RNA complexes through the earliest peptide catalysts to contemporary aminoacyl-tRNA synthetases. That scenario documents a path to increasing complexity that obviates the need for a single polymer to act both catalytically and as an informational molecule.
Figures







References
-
- Carter C.W., Jr. Cradles for Molecular Evolution. New Scientist. 1975 Mar 27;:784–787.
-
- Joyce G., Orgel L.E. Progress Toward Understanding the Origin of the RNA World. In: Gesteland R.F., Cech T.R., Atkins J., editors. The RNA World. 3rd ed. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 2006.
-
- Akst J. RNA World 2.0. The Scientist. 2014 Mar 1;:34–40.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

