Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates
- PMID: 25625844
- PMCID: PMC4623018
- DOI: 10.4161/15592294.2014.983381
Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates
Abstract
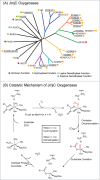
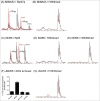
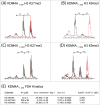
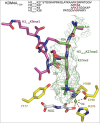
The JmjC-domain-containing 2-oxoglutarate-dependent oxygenases catalyze protein hydroxylation and N(ϵ)-methyllysine demethylation via hydroxylation. A subgroup of this family, the JmjC lysine demethylases (JmjC KDMs) are involved in histone modifications at multiple sites. There are conflicting reports as to the substrate selectivity of some JmjC oxygenases with respect to KDM activities. In this study, a panel of modified histone H3 peptides was tested for demethylation against 15 human JmjC-domain-containing proteins. The results largely confirmed known N(ϵ)-methyllysine substrates. However, the purified KDM4 catalytic domains showed greater substrate promiscuity than previously reported (i.e., KDM4A was observed to catalyze demethylation at H3K27 as well as H3K9/K36). Crystallographic analyses revealed that the N(ϵ)-methyllysine of an H3K27me3 peptide binds similarly to N(ϵ)-methyllysines of H3K9me3/H3K36me3 with KDM4A. A subgroup of JmjC proteins known to catalyze hydroxylation did not display demethylation activity. Overall, the results reveal that the catalytic domains of the KDM4 enzymes may be less selective than previously identified. They also draw a distinction between the N(ϵ)-methyllysine demethylation and hydroxylation activities within the JmjC subfamily. These results will be of use to those working on functional studies of the JmjC enzymes.
Keywords: 2OG oxygenases; 2OG, 2-oxoglutarate; Epigenetics; FIH, Factor Inhibiting HIF; H3, histone 3; HIF, Hypoxia Inducible Factor; JmjC histone demethylase; JmjC, Jumonji C-terminal; JmjN, Jumonji N-terminal; KDM, Lysine Demethylase; LSD, Lysine Specific Demethylase; MALDI-TOF MS, Matrix Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry; MINA53, Myc-Induced Nuclear Antigen with a molecular mass of 53 kDa; NO66, Nucleolar protein 66; PHD, Plant Homeodomain; Rp, Ribosomal protein; TPR, Tetratricopeptide repeat; demethylation; histone; methyllysine.
Figures
Similar articles
-
The catalytic domains of all human KDM5 JmjC demethylases catalyse N-methyl arginine demethylation.FEBS Lett. 2023 Apr;597(7):933-946. doi: 10.1002/1873-3468.14586. Epub 2023 Feb 7. FEBS Lett. 2023. PMID: 36700827 Free PMC article.
-
Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26.FEBS Lett. 2018 Oct;592(19):3264-3273. doi: 10.1002/1873-3468.13231. Epub 2018 Sep 14. FEBS Lett. 2018. PMID: 30156264 Free PMC article.
-
Lysine-241 Has a Role in Coupling 2OG Turnover with Substrate Oxidation During KDM4-Catalysed Histone Demethylation.Chembiochem. 2018 May 4;19(9):917-921. doi: 10.1002/cbic.201800002. Epub 2018 Apr 6. Chembiochem. 2018. PMID: 29443450 Free PMC article.
-
Structure-function relationships in KDM7 histone demethylases.Adv Protein Chem Struct Biol. 2019;117:113-125. doi: 10.1016/bs.apcsb.2019.08.005. Epub 2019 Sep 10. Adv Protein Chem Struct Biol. 2019. PMID: 31564306 Review.
-
Structure-function relationships of human JmjC oxygenases-demethylases versus hydroxylases.Curr Opin Struct Biol. 2016 Dec;41:62-72. doi: 10.1016/j.sbi.2016.05.013. Epub 2016 Jun 14. Curr Opin Struct Biol. 2016. PMID: 27309310 Review.
Cited by
-
Small molecule KDM4s inhibitors as anti-cancer agents.J Enzyme Inhib Med Chem. 2018 Dec;33(1):777-793. doi: 10.1080/14756366.2018.1455676. J Enzyme Inhib Med Chem. 2018. PMID: 29651880 Free PMC article.
-
Hole hopping through tyrosine/tryptophan chains protects proteins from oxidative damage.Proc Natl Acad Sci U S A. 2015 Sep 1;112(35):10920-5. doi: 10.1073/pnas.1512704112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195784 Free PMC article.
-
To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families.Int J Mol Sci. 2024 Jun 24;25(13):6900. doi: 10.3390/ijms25136900. Int J Mol Sci. 2024. PMID: 39000010 Free PMC article. Review.
-
The critical role of histone lysine demethylase KDM2B in cancer.Am J Transl Res. 2018 Aug 15;10(8):2222-2233. eCollection 2018. Am J Transl Res. 2018. PMID: 30210666 Free PMC article. Review.
-
The Lysine Demethylase dKDM2 Is Non-essential for Viability, but Regulates Circadian Rhythms in Drosophila.Front Genet. 2018 Sep 4;9:354. doi: 10.3389/fgene.2018.00354. eCollection 2018. Front Genet. 2018. PMID: 30233643 Free PMC article.
References
-
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13:343-57; PMID:22473383; http://dx.doi.org/10.1038/nrg3173 - DOI - PMC - PubMed
-
- Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol 2008; 4:152-6; PMID:18277970; http://dx.doi.org/10.1038/nchembio0308-152 - DOI - PubMed
-
- Walport LJ, Hopkinson RJ, Schofield CJ. Mechanisms of human histone and nucleic acid demethylases. Curr Opin Chem Biol 2012; 16:525-34; PMID:23063108; http://dx.doi.org/10.1016/j.cbpa.2012.09.015 - DOI - PubMed
-
- Ge W, Wolf A, Feng T, Ho CH, Sekirnik R, Zayer A, Granatino N, Cockman ME, Loenarz C, Loik ND, et al. . Oxygenase-catalyzed ribosome hydroxylation occurs in prokaryotes and humans. Nat Chem Biol 2012; 8:960-2; PMID:23103944; http://dx.doi.org/10.1038/nchembio.1093 - DOI - PMC - PubMed
-
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science 2007; 318:444-7; PMID:17947579; http://dx.doi.org/10.1126/science.1145801 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
