Mechanisms and pathways of innate immune activation and regulation in health and cancer
- PMID: 25625930
- PMCID: PMC4514086
- DOI: 10.4161/21645515.2014.979640
Mechanisms and pathways of innate immune activation and regulation in health and cancer
Abstract
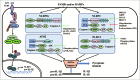
Research on innate immune signaling and regulation has recently focused on pathogen recognition receptors (PRRs) and their signaling pathways. Members of PRRs sense diverse microbial invasions or danger signals, and initiate innate immune signaling pathways, leading to proinflammatory cytokines production, which, in turn, instructs adaptive immune response development. Despite the diverse functions employed by innate immune signaling to respond to a variety of different pathogens, the innate immune response must be tightly regulated. Otherwise, aberrant, uncontrolled immune responses will lead to harmful, or even fatal, consequences. Therefore, it is essential to better discern innate immune signaling and many regulators, controlling various signaling pathways, have been identified. In this review, we focus on the recent advances in our understanding of the activation and regulation of innate immune signaling in the host response to pathogens and cancer.
Keywords: AIM2, absent in melanoma 2; ALRs, AIM2-like receptors; AMPK, AMP activated protein kinase; ASC, apoptosis-associated speck-like protein containing a CARD; Atg16L, autophagy related 16-like; BMM, bone marrow-derived macrophage; CARD, caspase recruitment domain; CDNs, cyclic dinucleotides; CLRs, C-type lectin receptors; CMV, cytomegalovirus; CYLD, the familial cylindromatosis tumor suppressor gene; DAMPs, danger-associated molecular patterns; DCs, dendritic cells; DDX41, DEAD (Asp-Glu-Ala-Asp) box polypeptide 41; ER, endoplasmic reticulum; GBP5, guanylate-binding protein 5; GSK3β, Glycogen synthase kinase 3β; HCC, hepatocellular carcinoma; IFI16, interferon, gamma-inducible protein 16; IFN, interferon; IKK, IkB kinase; IKKi, inducible IkB kinase; IRAK, interleukin-1 receptor-associated kinase; IRF, interferon regulatory factor; KSHV, Kaposi's sarcoma-associated herpesvirus; LBP, LPS-binding protein; LGP 2, laboratory of genetics and physiology 2; LPS, lipopolysaccharide; LRR, leucine-rich repeat; LT, lethal toxin; LUBAC, linear ubiquitin assembly complex; MAVS, mitochondrial antiviral signaling protein; MDA5, melanoma differentiation-associated protein 5; MDP, muramyl dipeptide; MIB, mind bomb; MyD88, myeloid differentiation factor 88; NAIPs, neuronal apoptosis inhibitory proteins; NEMO, NF-kB essential modulator; NLRs, Nod- like receptors; NOD, nucleotide-binding oligomerization domain; Nrdp1, neuregulin receptor degradation protein 1; PAMPs, pathogen-associated molecular patterns; PKC-d, protein kinase C delta; PKR, dsRNA-dependent protein kinase; PRRs; PRRs, pathogen recognition receptors; RACK1, receptor for activated C kinase 1; RAUL, RTA-associated E3 ligase; RIG-I, retinoic acid-inducible gene 1; RIP, receptor-interacting protein; RLRs, RIG-I-like receptors; ROS, reactive oxygen species; SARM, sterile a- and armadillo motif-containing protein; SIGIRR, single Ig IL-1-related receptor; SOCS, suppressor of cytokine signaling; STING, stimulator of interferon gene; TAK1, TGF-b-activating kinase 1; TANK, TRAF family-member-associated NF-kB activator; TBK1, TANK binding kinase 1; TIR, Toll IL-1 receptor; TIRAP, TIR domain-containing adapter protein; TLRs, Toll-like receptors; TRAF, TNFR-associated factor; TRAILR, tumor-necrosis factor-related apoptosis-inducing ligand receptor; TRAM, TRIF-related adaptor molecule; TRIF, TIR domain-containing adaptor inducing IFN-b; TRIMs, tripartite motif containing proteins; TRIP, TRAF-interacting protein; ULK1, autophagy related serine threonine UNC-51- like kinase; cDC, conventional dendritic cell; cGAS, cyclic GMP-AMP synthase; cIAP, cellular inhibitor of apoptosis protein; cancer; iE-DAP, g-D-glutamyl-meso-diaminopimelic acid; inflammation; innate immunity; pDC, plasmacytoid dendritic cell; type I interferon.
Figures


References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell 2006; 124:783-801; PMID:16497588; http://dx.doi.org/ 10.1016/j.cell.2006.02.015 - DOI - PubMed
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805-20; PMID:20303872; http://dx.doi.org/ 10.1016/j.cell.2010.01.022 - DOI - PubMed
-
- Paludan SR, Bowie AG. Immune sensing of DNA. Immunity 2013; 38:870-80; PMID:23706668; http://dx.doi.org/ 10.1016/j.immuni.2013.05.004 - DOI - PMC - PubMed
-
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol 2004; 5:987-95; PMID:15454922; http://dx.doi.org/ 10.1038/ni1112 - DOI - PubMed
-
- Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol 2005; 17:1-14; PMID:15585605; http://dx.doi.org/ 10.1093/intimm/dxh186 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
