Quantitative analysis of differences in copy numbers using read depth obtained from PCR-enriched samples and controls
- PMID: 25626454
- PMCID: PMC4384318
- DOI: 10.1186/s12859-014-0428-5
Quantitative analysis of differences in copy numbers using read depth obtained from PCR-enriched samples and controls
Abstract
Background: Next-generation sequencing (NGS) is rapidly becoming common practice in clinical diagnostics and cancer research. In addition to the detection of single nucleotide variants (SNVs), information on copy number variants (CNVs) is of great interest. Several algorithms exist to detect CNVs by analyzing whole genome sequencing data or data from samples enriched by hybridization-capture. PCR-enriched amplicon-sequencing data have special characteristics that have been taken into account by only one publicly available algorithm so far.
Results: We describe a new algorithm named quandico to detect copy number differences based on NGS data generated following PCR-enrichment. A weighted t-test statistic was applied to calculate probabilities (p-values) of copy number changes. We assessed the performance of the method using sequencing reads generated from reference DNA with known CNVs, and we were able to detect these variants with 98.6% sensitivity and 98.5% specificity which is significantly better than another recently described method for amplicon sequencing. The source code (R-package) of quandico is licensed under the GPLv3 and it is available at https://github.com/reineckef/quandico .
Conclusion: We demonstrated that our new algorithm is suitable to call copy number changes using data from PCR-enriched samples with high sensitivity and specificity even for single copy differences.
Figures
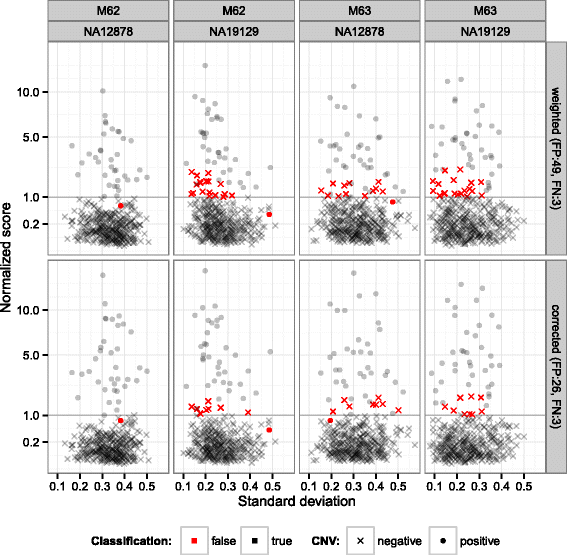
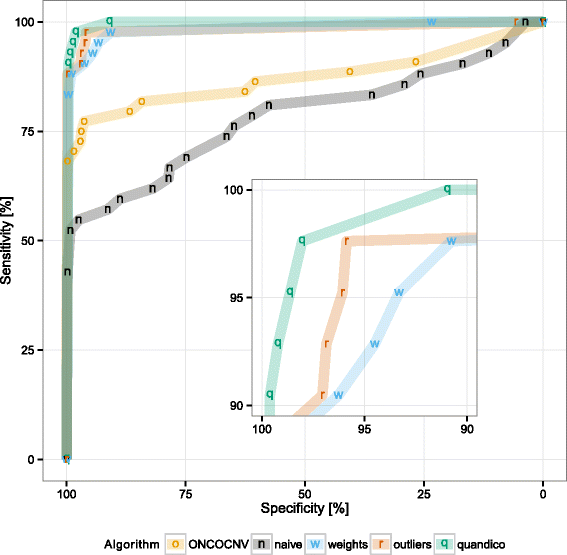
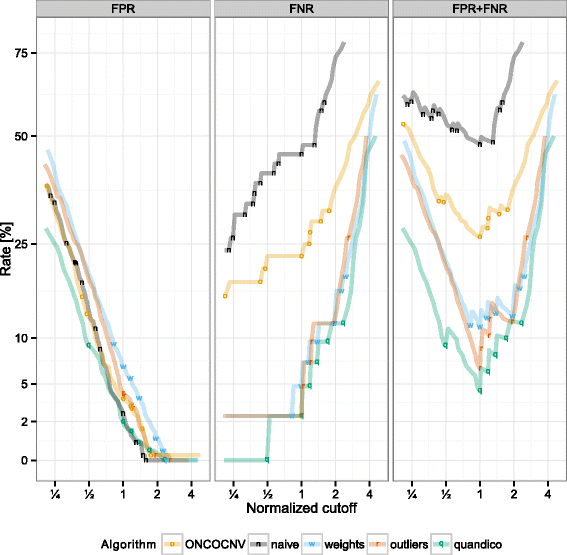
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

