MDAT- Aligning multiple domain arrangements
- PMID: 25626688
- PMCID: PMC4384290
- DOI: 10.1186/s12859-014-0442-7
MDAT- Aligning multiple domain arrangements
Abstract
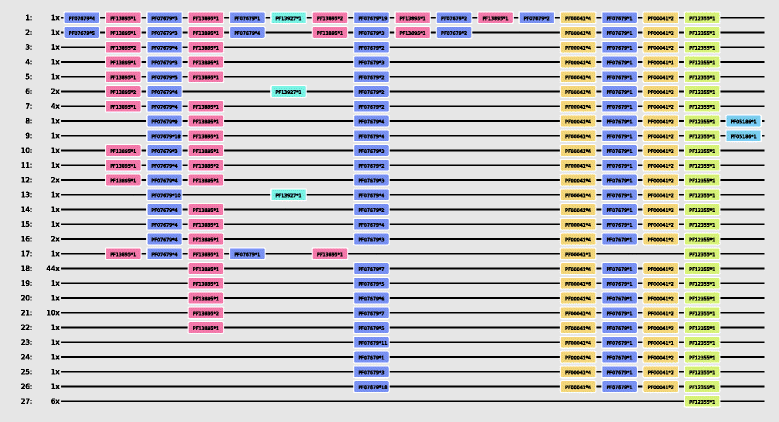
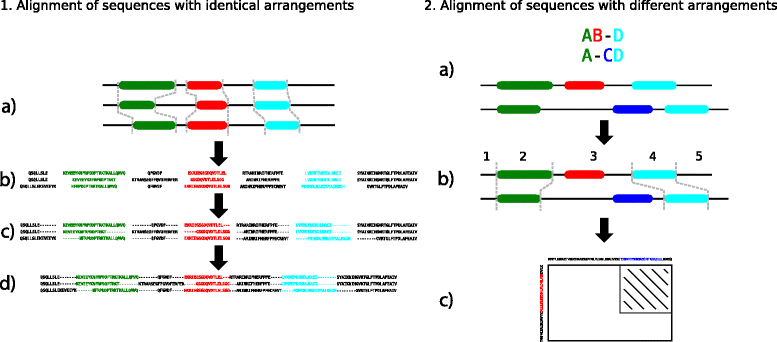
Background: Proteins are composed of domains, protein segments that fold independently from the rest of the protein and have a specific function. During evolution the arrangement of domains can change: domains are gained, lost or their order is rearranged. To facilitate the analysis of these changes we propose the use of multiple domain alignments.
Results: We developed an alignment program, called MDAT, which aligns multiple domain arrangements. MDAT extends earlier programs which perform pairwise alignments of domain arrangements. MDAT uses a domain similarity matrix to score domain pairs and aligns the domain arrangements using a consistency supported progressive alignment method.
Conclusion: MDAT will be useful for analysing changes in domain arrangements within and between protein families and will thus provide valuable insights into the evolution of proteins and their domains. MDAT is coded in C++, and the source code is freely available for download at http://www.bornberglab.org/pages/mdat .
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

