Putative E3 ubiquitin ligase of human rotavirus inhibits NF-κB activation by using molecular mimicry to target β-TrCP
- PMID: 25626907
- PMCID: PMC4324316
- DOI: 10.1128/mBio.02490-14
Putative E3 ubiquitin ligase of human rotavirus inhibits NF-κB activation by using molecular mimicry to target β-TrCP
Abstract
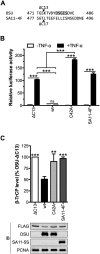
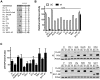
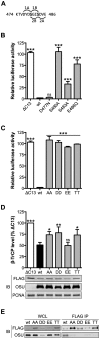
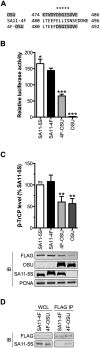
NF-κB plays a critical role in the induction and maintenance of innate and adaptive immune transcriptional programs. An associated inhibitor of κB protein (IκB) regulates NF-κB activation and contains a degron motif (DSGΦxS) that undergoes phosphorylation following pathogen recognition or other proinflammatory signals. The E3 ubiquitin ligase SCF(β-TrCP) recognizes this phosphodegron through its β-transducin repeat-containing protein (β-TrCP) subunit and induces IκB degradation, allowing NF-κB to translocate to the nucleus and modulate gene expression. Rotavirus (RV), a major cause of pediatric gastroenteritis, can block NF-κB activation through the action of its nonstructural protein NSP1, a putative E3 ubiquitin ligase that mediates the degradation of β-TrCP or other immunomodulatory proteins in a virus strain-specific manner. Here, we show that NSP1 targets β-TrCP by mimicking the IκB phosphodegron. The NSP1 proteins of most human and porcine RV strains conserve a C-terminal phosphodegron-like (PDL) motif, DSGΦS. Deletion of this motif or mutation of its serine residues disrupts NSP1-mediated degradation of β-TrCP and inhibition of NF-κB activation. Additionally, a point mutation within the phosphodegron-binding pocket protects β-TrCP from NSP1-mediated turnover. Fusion of the PDL motif to an NSP1 protein known to target other immunomodulatory proteins generates a chimeric NSP1 protein that can induce β-TrCP degradation and block NF-κB activation. Other viral proteins (Epstein-Barr virus LMP1, HIV-1 Vpu, and vaccinia virus A49) also contain a PDL motif and interact with β-TrCP to inhibit NF-κB activation. Taken together, these data suggest that targeting β-TrCP by molecular mimicry may be a common strategy used by human viruses to evade the host immune response.
Importance: The transcription factor NF-κB, a central regulator of the host response to infection, is a frequent target of viral antagonism. Pathogen detection activates NF-κB by inducing the phosphorylation of an associated inhibitor protein (IκB), which targets IκB for degradation by the E3 ubiquitin ligase β-TrCP. Rotavirus, a significant cause of childhood gastroenteritis, antagonizes NF-κB through the activity of its NSP1 protein, a putative E3 ubiquitin ligase that mediates β-TrCP turnover. Here, we show that NSP1 functions by mimicking the IκB phosphodegron recognized by β-TrCP. Nearly all human rotavirus strains conserve this motif at the NSP1 C terminus, and its removal disrupts NSP1 antagonist activity. This sequence conserves the biochemical properties of the IκB phosphodegron and can rescue antagonist activity when fused to an NSP1 protein otherwise inactive against β-TrCP. Other viral proteins also mimic IκB to disrupt NF-κB activation, indicating that this is an important immune evasion strategy.
Copyright © 2015 Morelli et al.
Figures


Similar articles
-
Rotavirus NSP1 Requires Casein Kinase II-Mediated Phosphorylation for Hijacking of Cullin-RING Ligases.mBio. 2017 Aug 29;8(4):e01213-17. doi: 10.1128/mBio.01213-17. mBio. 2017. PMID: 28851847 Free PMC article.
-
NSP1 of human rotaviruses commonly inhibits NF-κB signalling by inducing β-TrCP degradation.J Gen Virol. 2015 Jul;96(Pt 7):1768-76. doi: 10.1099/vir.0.000093. Epub 2015 Feb 20. J Gen Virol. 2015. PMID: 25701827
-
Rotavirus NSP1 inhibits NFkappaB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism.PLoS Pathog. 2009 Jan;5(1):e1000280. doi: 10.1371/journal.ppat.1000280. Epub 2009 Jan 30. PLoS Pathog. 2009. PMID: 19180189 Free PMC article.
-
Regulation of NF-κB by ubiquitination and degradation of the IκBs.Immunol Rev. 2012 Mar;246(1):77-94. doi: 10.1111/j.1600-065X.2012.01098.x. Immunol Rev. 2012. PMID: 22435548 Review.
-
Rotavirus and reovirus modulation of the interferon response.J Interferon Cytokine Res. 2009 Sep;29(9):559-67. doi: 10.1089/jir.2009.0072. J Interferon Cytokine Res. 2009. PMID: 19694545 Free PMC article. Review.
Cited by
-
Re-Examining Rotavirus Innate Immune Evasion: Potential Applications of the Reverse Genetics System.mBio. 2022 Aug 30;13(4):e0130822. doi: 10.1128/mbio.01308-22. Epub 2022 Jun 14. mBio. 2022. PMID: 35699371 Free PMC article. Review.
-
MHC class I expression in intestinal cells is reduced by rotavirus infection and increased in bystander cells lacking rotavirus antigen.Sci Rep. 2018 Jan 8;8(1):67. doi: 10.1038/s41598-017-18464-x. Sci Rep. 2018. PMID: 29311575 Free PMC article.
-
TRIM67 Suppresses TNFalpha-Triggered NF-kB Activation by Competitively Binding Beta-TrCP to IkBa.Front Immunol. 2022 Feb 22;13:793147. doi: 10.3389/fimmu.2022.793147. eCollection 2022. Front Immunol. 2022. PMID: 35273593 Free PMC article.
-
Differential dysregulation of β-TrCP1 and -2 by HIV-1 Vpu leads to inhibition of canonical and non-canonical NF-κB pathways in infected cells.mBio. 2023 Aug 31;14(4):e0329322. doi: 10.1128/mbio.03293-22. Epub 2023 Jun 21. mBio. 2023. PMID: 37341489 Free PMC article.
-
Viral Suppression of RIPK1-Mediated Signaling.mBio. 2021 Aug 31;12(4):e0172321. doi: 10.1128/mBio.01723-21. Epub 2021 Aug 10. mBio. 2021. PMID: 34372694 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

