Increased sample volume and use of quantitative reverse-transcription PCR can improve prediction of liver-to-blood inoculum size in controlled human malaria infection studies
- PMID: 25627033
- PMCID: PMC4318195
- DOI: 10.1186/s12936-015-0541-6
Increased sample volume and use of quantitative reverse-transcription PCR can improve prediction of liver-to-blood inoculum size in controlled human malaria infection studies
Abstract
Background: Controlled human malaria infection (CHMI) studies increasingly rely on nucleic acid test (NAT) methods to detect and quantify parasites in the blood of infected participants. The lower limits of detection and quantification vary amongst the assays used throughout the world, which may affect the ability of mathematical models to accurately estimate the liver-to-blood inoculum (LBI) values that are used to judge the efficacy of pre-erythrocytic vaccine and drug candidates.
Methods: Samples were collected around the time of onset of pre-patent parasitaemia from subjects who enrolled in two different CHMI clinical trials. Blood samples were tested for Plasmodium falciparum 18S rRNA and/or rDNA targets by different NAT methods and results were compared. Methods included an ultrasensitive, large volume modification of an established quantitative reverse transcription PCR (qRT-PCR) assay that achieves detection of as little as one parasite/mL of whole blood.
Results: Large volume qRT-PCR at the University of Washington was the most sensitive test and generated quantifiable data more often than any other NAT methodology. Standard quantitative PCR (qPCR) performed at the University of Oxford and standard volume qRT-PCR performed at the University of Washington were less sensitive than the large volume qRT-PCR, especially at 6.5 days after CHMI. In these trials, the proportion of participants for whom LBI could be accurately quantified using parasite density value greater than or equal to the lower limit of quantification was increased. A greater improvement would be expected in trials in which numerous subjects receive a lower LBI or low dose challenge.
Conclusions: Standard qPCR and qRT-PCR methods with analytical sensitivities of ~20 parasites/mL probably suffice for most CHMI purposes, but the newly developed large volume qRT-PCR may be able to answer specific questions when more analytical sensitivity is required.
Figures

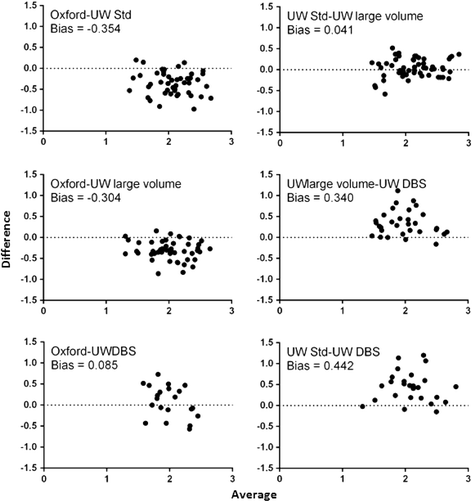
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

