The principal mRNA nuclear export factor NXF1:NXT1 forms a symmetric binding platform that facilitates export of retroviral CTE-RNA
- PMID: 25628361
- PMCID: PMC4330390
- DOI: 10.1093/nar/gkv032
The principal mRNA nuclear export factor NXF1:NXT1 forms a symmetric binding platform that facilitates export of retroviral CTE-RNA
Abstract
The NXF1:NXT1 complex (also known as TAP:p15) is a general mRNA nuclear export factor that is conserved from yeast to humans. NXF1 is a modular protein constructed from four domains (RRM, LRR, NTF2-like and UBA domains). It is currently unclear how NXF1:NXT1 binds transcripts and whether there is higher organization of the NXF1 domains. We report here the 3.4 Å resolution crystal structure of the first three domains of human NXF1 together with NXT1 that has two copies of the complex in the asymmetric unit arranged to form an intimate domain-swapped dimer. In this dimer, the linkers between the NXF1 LRR and NTF2-like domains interact with NXT1, generating a 2-fold symmetric platform in which the RNA-binding RRM, LRR and NTF2-like domains are arranged on one face. In addition to bulk transcripts, NXF1:NXT1 also facilitates the export of unspliced retroviral genomic RNA from simple type-D retroviruses such as SRV-1 that contain a constitutive transport element (CTE), a cis-acting 2-fold symmetric RNA stem-loop motif. Complementary structural, biochemical and cellular techniques indicated that the formation of a symmetric RNA binding platform generated by dimerization of NXF1:NXT1 facilitates the recognition of CTE-RNA and promotes its nuclear export.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
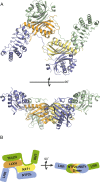





References
-
- Niño C.A., Hérissant L, Babour A., Dargemont C. mRNA nuclear export in yeast. Chem. Rev. 2013;113:8523–8545. - PubMed
-
- Stewart M. Nuclear export of mRNA. Trends Biochem. Sci. 2010;35:609–617. - PubMed
-
- Grüter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B.K., Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell. 1998;1:649–659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

