Regulation of mammalian nucleotide metabolism and biosynthesis
- PMID: 25628363
- PMCID: PMC4344498
- DOI: 10.1093/nar/gkv047
Regulation of mammalian nucleotide metabolism and biosynthesis
Abstract
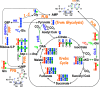
Nucleotides are required for a wide variety of biological processes and are constantly synthesized de novo in all cells. When cells proliferate, increased nucleotide synthesis is necessary for DNA replication and for RNA production to support protein synthesis at different stages of the cell cycle, during which these events are regulated at multiple levels. Therefore the synthesis of the precursor nucleotides is also strongly regulated at multiple levels. Nucleotide synthesis is an energy intensive process that uses multiple metabolic pathways across different cell compartments and several sources of carbon and nitrogen. The processes are regulated at the transcription level by a set of master transcription factors but also at the enzyme level by allosteric regulation and feedback inhibition. Here we review the cellular demands of nucleotide biosynthesis, their metabolic pathways and mechanisms of regulation during the cell cycle. The use of stable isotope tracers for delineating the biosynthetic routes of the multiple intersecting pathways and how these are quantitatively controlled under different conditions is also highlighted. Moreover, the importance of nucleotide synthesis for cell viability is discussed and how this may lead to potential new approaches to drug development in diseases such as cancer.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


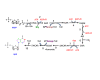
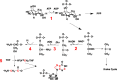
 ) are incorporated into the ribose unit (via PPP), uracil ring (via the Krebs cycle–pyrimidine synthesis path or PYR) of UMP or adenine ring (via the one-carbon or 1-C to purine synthesis path or PUR) of AMP (structures shown). The 13C (
) are incorporated into the ribose unit (via PPP), uracil ring (via the Krebs cycle–pyrimidine synthesis path or PYR) of UMP or adenine ring (via the one-carbon or 1-C to purine synthesis path or PUR) of AMP (structures shown). The 13C ( ) and 15N labels (
) and 15N labels ( ) from 13C5,15N2-Gln are expected to go into the uracil ring (via the anaplerotic glutaminolysis or GLS-Krebs cycle-PYR path) of UMP and the adenine ring (via the PUR path) of AMP. The color of the label for atomic positions in the UMP and AMP structures is matched with that of 13C or 15N label derived from the glucose or glutamine tracer, except for C4-C6 of UMP where glucose or Gln-derived 13C is not delineated. Three examples of labeled uracil ring delineate contribution of 13C from 13C6-Glc or 13C5,15N2-Gln after one Krebs cycle turn without or with pyruvate carboxylation. The 13C labeling patterns of the Krebs cycle intermediates and Asp account for the 13C scrambling in succinate due to its symmetry and anaplerotic input (green arrows and
) from 13C5,15N2-Gln are expected to go into the uracil ring (via the anaplerotic glutaminolysis or GLS-Krebs cycle-PYR path) of UMP and the adenine ring (via the PUR path) of AMP. The color of the label for atomic positions in the UMP and AMP structures is matched with that of 13C or 15N label derived from the glucose or glutamine tracer, except for C4-C6 of UMP where glucose or Gln-derived 13C is not delineated. Three examples of labeled uracil ring delineate contribution of 13C from 13C6-Glc or 13C5,15N2-Gln after one Krebs cycle turn without or with pyruvate carboxylation. The 13C labeling patterns of the Krebs cycle intermediates and Asp account for the 13C scrambling in succinate due to its symmetry and anaplerotic input (green arrows and  ) from pyruvate carboxylation into the Krebs cycle after the first turn. Open circles: 12C; HK: hexokinase; G6PDH: glucose-6-phosphate dehydrogenase; PDH: pyruvate dehydrogenase; GLS: glutaminase; PCB: pyruvate carboxylase; OAA: oxaloacetate; αKG: α-ketoglutarate; exo: exocyclic.
) from pyruvate carboxylation into the Krebs cycle after the first turn. Open circles: 12C; HK: hexokinase; G6PDH: glucose-6-phosphate dehydrogenase; PDH: pyruvate dehydrogenase; GLS: glutaminase; PCB: pyruvate carboxylase; OAA: oxaloacetate; αKG: α-ketoglutarate; exo: exocyclic.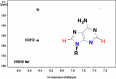
References
-
- Sigoillot F.D., Berkowski J.A., Sigoillot S.M., Kotsis D.H., Guy H.I. Cell cycle-dependent regulation of pyrimidine biosynthesis. J. Biol. Chem. 2003;278:3403–3409. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

