The family of LSU-like proteins
- PMID: 25628631
- PMCID: PMC4292543
- DOI: 10.3389/fpls.2014.00774
The family of LSU-like proteins
Abstract
The plant response to sulfur deficiency includes extensive metabolic changes which can be monitored at various levels (transcriptome, proteome, metabolome) even before the first visible symptoms of sulfur starvation appear. Four members of the plant-specific LSU (response to Low SUlfur) gene family occur in Arabidopsis thaliana (LSU1-4). Variable numbers of LSU genes occur in other plant species but they were studied only in Arabidopsis and tobacco. Three out of four of the Arabidopsis LSU genes are induced by sulfur deficiency. The LSU-like genes in tobacco were characterized as UP9 (UPregulated by sulfur deficit 9). LSU-like proteins do not have characteristic domains that provide clues to their function. Despite having only moderate primary sequence conservation they share several common features including small size, a coiled-coil secondary structure and short conserved motifs in specific positions. Although the precise function of LSU-like proteins is still unknown there is some evidence that members of the LSU family are involved in plant responses to environmental challenges, such as sulfur deficiency, and possibly in plant immune responses. Various bioinformatic approaches have identified LSU-like proteins as important hubs for integration of signals from environmental stimuli. In this paper we review a variety of published data on LSU gene expression, the properties of lsu mutants and features of LSU-like proteins in the hope of shedding some light on their possible role in plant metabolism.
Keywords: Arabidopsis; OAS; SALK mutants; UP9; coiled coil; ethylene; gene expression; tobacco.
Figures



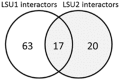
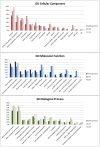
References
-
- Adie B., Chico J. M., Rubio-Somoza I., Solano R. (2007). Modulation of plant defenses by ethylene. J. Plant Growth Regul. 26 160–177 10.1007/s00344-007-0012-6 - DOI
-
- Ascencio-Ibanez J. T., Sozzani R., Lee T. J., Chu T. M., Wolfinger R. D., Cella R., et al. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148 436–454 10.1104/pp.108.121038 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

