Impact of viral activators and epigenetic regulators on HIV-1 LTRs containing naturally occurring single nucleotide polymorphisms
- PMID: 25629043
- PMCID: PMC4299542
- DOI: 10.1155/2015/320642
Impact of viral activators and epigenetic regulators on HIV-1 LTRs containing naturally occurring single nucleotide polymorphisms
Abstract
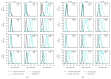
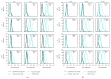
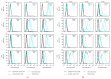
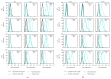
Following human immunodeficiency virus type 1 (HIV-1) integration into host cell DNA, the viral promoter can become transcriptionally silent in the absence of appropriate signals and factors. HIV-1 gene expression is dependent on regulatory elements contained within the long terminal repeat (LTR) that drive the synthesis of viral RNAs and proteins through interaction with multiple host and viral factors. Previous studies identified single nucleotide polymorphisms (SNPs) within CCAAT/enhancer binding protein (C/EBP) site I and Sp site III (3T, C-to-T change at position 3, and 5T, C-to-T change at position 5 of the binding site, respectively, when compared to the consensus B sequence) that are low affinity binding sites and correlate with more advanced stages of HIV-1 disease. Stably transfected cell lines containing the wild type, 3T, 5T, and 3T5T LTRs were developed utilizing bone marrow progenitor, T, and monocytic cell lines to explore the LTR phenotypes associated with these genotypic changes from an integrated chromatin-based microenvironment. Results suggest that in nonexpressing cell clones LTR-driven gene expression occurs in a SNP-specific manner in response to LTR activation or treatment with trichostatin A treatment, indicating a possible cell type and SNP-specific mechanism behind the epigenetic control of LTR activation.
Figures
Similar articles
-
Functional properties of the HIV-1 long terminal repeat containing single-nucleotide polymorphisms in Sp site III and CCAAT/enhancer binding protein site I.Virol J. 2014 May 16;11:92. doi: 10.1186/1743-422X-11-92. Virol J. 2014. PMID: 24886416 Free PMC article.
-
Analysis of the HIV-1 LTR NF-kappaB-proximal Sp site III: evidence for cell type-specific gene regulation and viral replication.Virology. 2000 Sep 1;274(2):262-77. doi: 10.1006/viro.2000.0476. Virology. 2000. PMID: 10964770
-
Structural and functional evolution of human immunodeficiency virus type 1 long terminal repeat CCAAT/enhancer binding protein sites and their use as molecular markers for central nervous system disease progression.J Neurovirol. 2003 Feb;9(1):55-68. doi: 10.1080/13550280390173292. J Neurovirol. 2003. PMID: 12587069
-
Inhibitors of HIV-1 transcription.Trends Microbiol. 1994 May;2(5):164-9. doi: 10.1016/0966-842x(94)90666-1. Trends Microbiol. 1994. PMID: 8055180 Review.
-
HIV UTR, LTR, and Epigenetic Immunity.Viruses. 2022 May 18;14(5):1084. doi: 10.3390/v14051084. Viruses. 2022. PMID: 35632825 Free PMC article. Review.
Cited by
-
Computational Analysis Concerning the Impact of DNA Accessibility on CRISPR-Cas9 Cleavage Efficiency.Mol Ther. 2020 Jan 8;28(1):19-28. doi: 10.1016/j.ymthe.2019.10.008. Epub 2019 Oct 15. Mol Ther. 2020. PMID: 31672284 Free PMC article.
-
HIV-1 Promoter Single Nucleotide Polymorphisms Are Associated with Clinical Disease Severity.PLoS One. 2016 Apr 21;11(4):e0150835. doi: 10.1371/journal.pone.0150835. eCollection 2016. PLoS One. 2016. PMID: 27100290 Free PMC article.
-
Diverse fates of uracilated HIV-1 DNA during infection of myeloid lineage cells.Elife. 2016 Sep 20;5:e18447. doi: 10.7554/eLife.18447. Elife. 2016. PMID: 27644592 Free PMC article.
References
-
- Fischer-Smith T., Croul S., Adeniyi A., et al. Macrophage/microglial accumulation and proliferating cell nuclear antigen expression in the central nervous system in human immunodeficiency virus encephalopathy. The American Journal of Pathology. 2004;164(6):2089–2099. doi: 10.1016/S0002-9440(10)63767-4. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

