PCR-activated cell sorting for cultivation-free enrichment and sequencing of rare microbes
- PMID: 25629401
- PMCID: PMC4309575
- DOI: 10.1371/journal.pone.0113549
PCR-activated cell sorting for cultivation-free enrichment and sequencing of rare microbes
Abstract
Microbial systems often exhibit staggering diversity, making the study of rare, interesting species challenging. For example, metagenomic analyses of mixed-cell populations are often dominated by the sequences of the most abundant organisms, while those of rare microbes are detected only at low levels, if at all. To overcome this, selective cultivation or fluorescence-activated cell sorting (FACS) can be used to enrich for the target species prior to sequence analysis; however, since most microbes cannot be grown in the lab, cultivation strategies often fail, while cell sorting requires techniques to uniquely label the cell type of interest, which is often not possible with uncultivable microbes. Here, we introduce a culture-independent strategy for sorting microbial cells based on genomic content, which we term PCR-activated cell sorting (PACS). This technology, which utilizes the power of droplet-based microfluidics, is similar to FACS in that it uses a fluorescent signal to uniquely identify and sort target species. However, PACS differs importantly from FACS in that the signal is generated by performing PCR assays on the cells in microfluidic droplets, allowing target cells to be identified with high specificity with suitable design of PCR primers and TaqMan probes. The PACS assay is general, requires minimal optimization and, unlike antibody methods, can be developed without access to microbial antigens. Compared to non-specific methods in which cells are sorted based on size, granularity, or the ability to take up dye, PACS enables genetic sequence-specific sorting and recovery of the cell genomes. In addition to sorting microbes, PACS can be applied to eukaryotic cells, viruses, and naked nucleic acids.
Conflict of interest statement
Figures
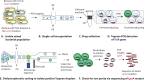
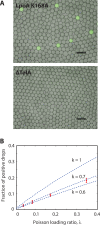
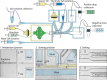
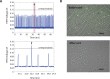
Similar articles
-
Peering below the diffraction limit: robust and specific sorting of viruses with flow cytometry.Virol J. 2016 Dec 1;13(1):201. doi: 10.1186/s12985-016-0655-7. Virol J. 2016. PMID: 27906039 Free PMC article.
-
RNA-Seq following PCR-based sorting reveals rare cell transcriptional signatures.BMC Genomics. 2016 May 17;17:361. doi: 10.1186/s12864-016-2694-2. BMC Genomics. 2016. PMID: 27189161 Free PMC article.
-
Optofluidic Raman-activated cell sorting for targeted genome retrieval or cultivation of microbial cells with specific functions.Nat Protoc. 2021 Feb;16(2):634-676. doi: 10.1038/s41596-020-00427-8. Epub 2020 Dec 11. Nat Protoc. 2021. PMID: 33311714
-
Go with the flow or solitary confinement: a look inside the single-cell toolbox for isolation of rare and uncultured microbes.Curr Opin Microbiol. 2018 Aug;44:1-8. doi: 10.1016/j.mib.2018.05.002. Epub 2018 Jun 13. Curr Opin Microbiol. 2018. PMID: 29908491 Review.
-
Developments in label-free microfluidic methods for single-cell analysis and sorting.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019 Jan;11(1):e1529. doi: 10.1002/wnan.1529. Epub 2018 Apr 24. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2019. PMID: 29687965 Free PMC article. Review.
Cited by
-
Single-Cell Microfluidics: A Primer for Microbiologists.J Phys Chem B. 2024 Oct 24;128(42):10311-10328. doi: 10.1021/acs.jpcb.4c02746. Epub 2024 Oct 14. J Phys Chem B. 2024. PMID: 39400277 Free PMC article. Review.
-
DEPICT-seq: Single-Cell Transcriptomic Analysis of Rare Cell Subsets Isolated via Nucleic Acid Cytometry.Anal Chem. 2024 Oct 15;96(41):16236-16243. doi: 10.1021/acs.analchem.4c03075. Epub 2024 Sep 17. Anal Chem. 2024. PMID: 39287475 Free PMC article.
-
Discovery of new protein families and functions: new challenges in functional metagenomics for biotechnologies and microbial ecology.Front Microbiol. 2015 Jun 5;6:563. doi: 10.3389/fmicb.2015.00563. eCollection 2015. Front Microbiol. 2015. PMID: 26097471 Free PMC article. Review.
-
Rapid parallel generation of a fluorescently barcoded drop library from a microtiter plate using the plate-interfacing parallel encapsulation (PIPE) chip.Lab Chip. 2022 Nov 22;22(23):4735-4745. doi: 10.1039/d2lc00909a. Lab Chip. 2022. PMID: 36367139 Free PMC article.
-
Review: Microbial analysis in dielectrophoretic microfluidic systems.Anal Chim Acta. 2017 May 8;966:11-33. doi: 10.1016/j.aca.2017.02.024. Epub 2017 Mar 6. Anal Chim Acta. 2017. PMID: 28372723 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

