Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway
- PMID: 25629602
- PMCID: PMC4317492
- DOI: 10.1038/ncomms7078
Temporal and spatial regulation of translation in the mammalian oocyte via the mTOR-eIF4F pathway
Abstract
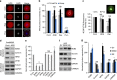
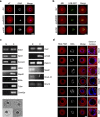
The fully grown mammalian oocyte is transcriptionally quiescent and utilizes only transcripts synthesized and stored during early development. However, we find that an abundant RNA population is retained in the oocyte nucleus and contains specific mRNAs important for meiotic progression. Here we show that during the first meiotic division, shortly after nuclear envelope breakdown, translational hotspots develop in the chromosomal area and in a region that was previously surrounded the nucleus. These distinct translational hotspots are separated by endoplasmic reticulum and Lamin, and disappear following polar body extrusion. Chromosomal translational hotspots are controlled by the activity of the mTOR-eIF4F pathway. Here we reveal a mechanism that-following the resumption of meiosis-controls the temporal and spatial translation of a specific set of transcripts required for normal spindle assembly, chromosome alignment and segregation.
Figures
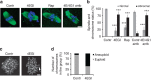
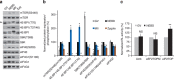
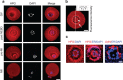
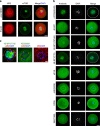

References
-
- Curtis D., Lehmann R. & Zamore P. D. Translational regulation in development. Cell 81, 171–178 (1995). - PubMed
-
- Raff R. A., Colot H. V., Selvig S. E. & Gross P. R. Oogenetic origin of messenger RNA for embryonic synthesis of microtubule proteins. Nature 235, 211–214 (1972). - PubMed
-
- Capco D. G. & Jeffery W. R. Origin and spatial distribution of maternal messenger RNA during oogenesis of an insect, Oncopeltus fasciatus. J. Cell Sci. 39, 63–76 (1979). - PubMed
-
- De La Fuente R. et al.. Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev. Biol. 275, 447–458 (2004). - PubMed
-
- Brandhorst B. P. Informational content of the echinoderm egg. Dev. Biol. (NY) 1, 525–576 (1985). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

