ST-246 is a key antiviral to inhibit the viral F13L phospholipase, one of the essential proteins for orthopoxvirus wrapping
- PMID: 25630650
- PMCID: PMC7539645
- DOI: 10.1093/jac/dku545
ST-246 is a key antiviral to inhibit the viral F13L phospholipase, one of the essential proteins for orthopoxvirus wrapping
Abstract
Objectives: ST-246 is one of the key antivirals being developed to fight orthopoxvirus (OPV) infections. Its exact mode of action is not completely understood, but it has been reported to interfere with the wrapping of infectious virions, for which F13L (peripheral membrane protein) and B5R (type I glycoprotein) are required. Here we monitored the appearance of ST-246 resistance to identify its molecular target.
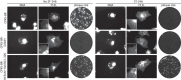
Methods: Vaccinia virus (VACV), cowpox virus (CPXV) and camelpox virus (CMLV) with reduced susceptibility to ST-246 were selected in cell culture and further characterized by antiviral assays and immunofluorescence. A panel of recombinant OPVs was engineered and a putative 3D model of F13L coupled with molecular docking was used to visualize drug-target interaction. The F13L gene of 65 CPXVs was sequenced to investigate F13L amino acid heterogeneity.
Results: Amino acid substitutions or insertions were found in the F13L gene of six drug-resistant OPVs and production of four F13L-recombinant viruses confirmed their role(s) in the occurrence of ST-246 resistance. F13L, but not B5R, knockout OPVs showed resistance to ST-246. ST-246 treatment of WT OPVs delocalized F13L- and B5R-encoded proteins and blocked virus wrapping. Putative modelling of F13L and ST-246 revealed a probable pocket into which ST-246 penetrates. None of the identified amino acid changes occurred naturally among newly sequenced or NCBI-derived OPV F13L sequences.
Conclusions: Besides demonstrating that F13L is a direct target of ST-246, we also identified novel F13L residues involved in the interaction with ST-246. These findings are important for ST-246 use in the clinic and crucial for future drug-resistance surveillance programmes.
Keywords: antivirals; drug resistance; mechanisms of action; poxvirus; vaccinia virus.
© The Author 2015. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures






References
-
- Haase O, Moser A, Rose C, et al. Generalized cowpox infection in a patient with Darier disease. Br J Dermatol 2011; 164: 1116–8. - PubMed
-
- Vogel S, Sardy M, Glos K, et al. The Munich outbreak of cutaneous cowpox infection: transmission by infected pet rats. Acta Derm Venereol 2012; 92: 126–31. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

