The origin of the ADAR gene family and animal RNA editing
- PMID: 25630791
- PMCID: PMC4323055
- DOI: 10.1186/s12862-015-0279-3
The origin of the ADAR gene family and animal RNA editing
Abstract
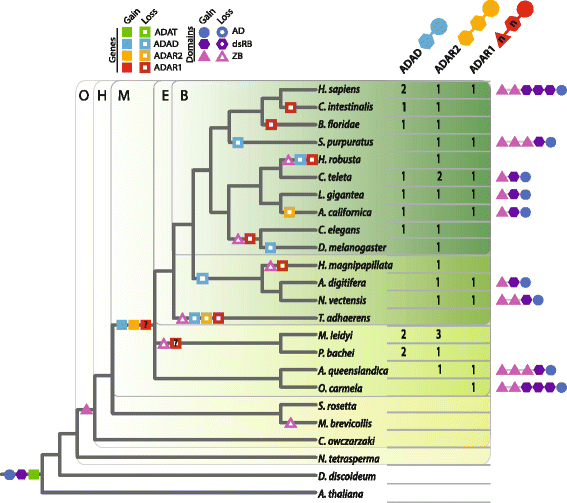
Background: ADAR (adenosine deaminase acting on RNA) proteins convert adenosine into inosine in double-stranded RNAs and have been shown to increase gene product diversity in a number of bilaterians, particularly mammals and flies. This enzyme family appears to have evolved from an ADAT (adenosine deaminase acting on tRNA) ancestor, via the addition of a double-stranded RNA binding domain. The modern vertebrate ADAR family is comprised of ADAD, ADAR2 and ADAR1, each of which has a conserved domain architecture. To reconstruct the origin of this protein family, we identified and categorised ADAR family members encoded in the genomes and/or transcriptomes of early-branching metazoan and closely related non-metazoan taxa, including thirteen sponge and ten ctenophore species.
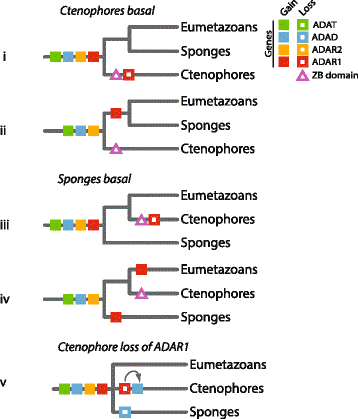
Results: We demonstrate that the ADAR protein family is a metazoan innovation, with the three ADAR subtypes being present in representatives of the earliest phyletic lineages of animals - sponges and ctenophores - but not in other closely related choanoflagellate and filasterean holozoans. ADAR1 is missing from all ctenophore genomes and transcriptomes surveyed. Depending on the relationship of sponges and ctenophores to the rest of the Metazoa, this is consistent with either ADAR1 being lost in ctenophores, as it has been in multiple metazoan lineages, or being an innovation that evolved after ctenophores diverged from the rest of the animal kingdom. The presence of Z-DNA binding domains in some sponge ADARs indicates an ancestral ADAR included this domain and it has been lost in multiple animal lineages.
Conclusions: The ADAR family appears to be a metazoan innovation, with all family members in place in the earliest phyletic branches of the crown Metazoa. The presence of ADARs in sponges and ctenophores is consistent with A-to-I editing being a post-transcriptional regulatory mechanism that was used by the last common ancestor to all living animals and subsequently has been preserved in most modern lineages.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

