The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population
- PMID: 25631317
- PMCID: PMC4391556
- DOI: 10.1534/genetics.115.174664
The Drosophila genome nexus: a population genomic resource of 623 Drosophila melanogaster genomes, including 197 from a single ancestral range population
Abstract
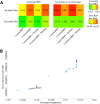
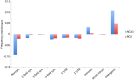
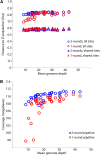
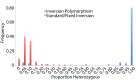
Hundreds of wild-derived Drosophila melanogaster genomes have been published, but rigorous comparisons across data sets are precluded by differences in alignment methodology. The most common approach to reference-based genome assembly is a single round of alignment followed by quality filtering and variant detection. We evaluated variations and extensions of this approach and settled on an assembly strategy that utilizes two alignment programs and incorporates both substitutions and short indels to construct an updated reference for a second round of mapping prior to final variant detection. Utilizing this approach, we reassembled published D. melanogaster population genomic data sets and added unpublished genomes from several sub-Saharan populations. Most notably, we present aligned data from phase 3 of the Drosophila Population Genomics Project (DPGP3), which provides 197 genomes from a single ancestral range population of D. melanogaster (from Zambia). The large sample size, high genetic diversity, and potentially simpler demographic history of the DPGP3 sample will make this a highly valuable resource for fundamental population genetic research. The complete set of assemblies described here, termed the Drosophila Genome Nexus, presently comprises 623 consistently aligned genomes and is publicly available in multiple formats with supporting documentation and bioinformatic tools. This resource will greatly facilitate population genomic analysis in this model species by reducing the methodological differences between data sets.
Keywords: Drosophila Genome Nexus; Drosophila melanogaster; genome assembly; population genomics.
Copyright © 2015 by the Genetics Society of America.
Figures

References
-
- Aulard S., David J. R., Lemeunier F., 2002. Chromosomal inversion polymorphism in Afrotropical populations of Drosophila melanogaster. Genet. Res. 79: 49–63. - PubMed
-
- Baudry E., Vinier B., Veuille M., 2004. Non-African populations of Drosophila melanogaster have a unique origin. Mol. Biol. Evol. 21: 1482–1491. - PubMed
-
- Begun D. J., Aquadro C. F., 1993. African and North American populations of Drosophila melanogaster are very different at the DNA level. Nature 365: 548–550. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

