13C NMR detects conformational change in the 100-kD membrane transporter ClC-ec1
- PMID: 25631353
- PMCID: PMC4398623
- DOI: 10.1007/s10858-015-9898-7
13C NMR detects conformational change in the 100-kD membrane transporter ClC-ec1
Abstract
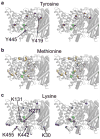
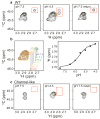
CLC transporters catalyze the exchange of Cl(-) for H(+) across cellular membranes. To do so, they must couple Cl(-) and H(+) binding and unbinding to protein conformational change. However, the sole conformational changes distinguished crystallographically are small movements of a glutamate side chain that locally gates the ion-transport pathways. Therefore, our understanding of whether and how global protein dynamics contribute to the exchange mechanism has been severely limited. To overcome the limitations of crystallography, we used solution-state (13)C-methyl NMR with labels on methionine, lysine, and engineered cysteine residues to investigate substrate (H(+)) dependent conformational change outside the restraints of crystallization. We show that methyl labels in several regions report H(+)-dependent spectral changes. We identify one of these regions as Helix R, a helix that extends from the center of the protein, where it forms the part of the inner gate to the Cl(-)-permeation pathway, to the extracellular solution. The H(+)-dependent spectral change does not occur when a label is positioned just beyond Helix R, on the unstructured C-terminus of the protein. Together, the results suggest that H(+) binding is mechanistically coupled to closing of the intracellular access-pathway for Cl(-).
Figures





References
-
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. - PubMed
-
- Accardi A, Lobet S, Williams C, Miller C, Dutzler R. Synergism between halide binding and proton transport in a CLC-type exchanger. J Mol Biol. 2006;362:691–699. - PubMed
-
- Accardi A, Miller C. Secondary active transport mediated by a prokaryotic homologue of ClC Cl- channels. Nature. 2004;427:803–807. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

