Systemic infection generates a local-like immune response of the bacteriome organ in insect symbiosis
- PMID: 25632977
- PMCID: PMC6738764
- DOI: 10.1159/000368928
Systemic infection generates a local-like immune response of the bacteriome organ in insect symbiosis
Abstract
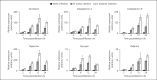
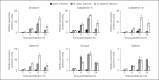
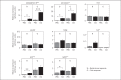
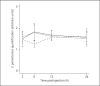
Endosymbiosis is common in insects thriving in nutritionally unbalanced habitats. The cereal weevil, Sitophilus oryzae, houses Sodalis pierantonius, a Gram-negative intracellular symbiotic bacterium (endosymbiont), within a dedicated organ called a bacteriome. Recent data have shown that the bacteriome expresses certain immune genes that result in local symbiont tolerance and control. Here, we address the question of whether and how the bacteriome responds to insect infections involving exogenous bacteria. We have established an infection model by challenging weevil larvae with the Gram-negative bacterium Dickeya dadantii. We showed that D. dadantii infects host tissues and triggers a systemic immune response. Gene transcript analysis indicated that the bacteriome is also immune responsive, but it expresses immune effector genes to a lesser extent than the systemic and intestinal responses. Most genes putatively involved in immune pathways remain weakly expressed in the bacteriome following D. dadantii infection. Moreover, quantitative PCR experiments showed that the endosymbiont load is not affected by insect infection or the resulting bacteriome immune activation. Thus, the contained immune effector gene expression in the bacteriome may prevent potentially harmful effects of the immune response on endosymbionts, whilst efficiently protecting them from bacterial intruders.
© 2015 S. Karger AG, Basel.
Figures


Similar articles
-
What can a weevil teach a fly, and reciprocally? Interaction of host immune systems with endosymbionts in Glossina and Sitophilus.BMC Microbiol. 2018 Nov 23;18(Suppl 1):150. doi: 10.1186/s12866-018-1278-5. BMC Microbiol. 2018. PMID: 30470176 Free PMC article. Review.
-
An IMD-like pathway mediates both endosymbiont control and host immunity in the cereal weevil Sitophilus spp.Microbiome. 2018 Jan 8;6(1):6. doi: 10.1186/s40168-017-0397-9. Microbiome. 2018. PMID: 29310713 Free PMC article.
-
Host gene response to endosymbiont and pathogen in the cereal weevil Sitophilus oryzae.BMC Microbiol. 2012 Jan 18;12 Suppl 1(Suppl 1):S14. doi: 10.1186/1471-2180-12-S1-S14. BMC Microbiol. 2012. PMID: 22375912 Free PMC article.
-
Identification of the weevil immune genes and their expression in the bacteriome tissue.BMC Biol. 2008 Oct 16;6:43. doi: 10.1186/1741-7007-6-43. BMC Biol. 2008. PMID: 18925938 Free PMC article.
-
Insect immune system maintains long-term resident bacteria through a local response.J Insect Physiol. 2013 Feb;59(2):232-9. doi: 10.1016/j.jinsphys.2012.06.015. Epub 2012 Jul 4. J Insect Physiol. 2013. PMID: 22771302 Review.
Cited by
-
Characterization of Antibacterial Activities of Eastern Subterranean Termite, Reticulitermes flavipes, against Human Pathogens.PLoS One. 2016 Sep 9;11(9):e0162249. doi: 10.1371/journal.pone.0162249. eCollection 2016. PLoS One. 2016. PMID: 27611223 Free PMC article.
-
What can a weevil teach a fly, and reciprocally? Interaction of host immune systems with endosymbionts in Glossina and Sitophilus.BMC Microbiol. 2018 Nov 23;18(Suppl 1):150. doi: 10.1186/s12866-018-1278-5. BMC Microbiol. 2018. PMID: 30470176 Free PMC article. Review.
-
Bacterial cell biology outside the streetlight.Environ Microbiol. 2016 Sep;18(8):2305-18. doi: 10.1111/1462-2920.13406. Epub 2016 Jul 15. Environ Microbiol. 2016. PMID: 27306428 Free PMC article. Review.
-
Efficient compartmentalization in insect bacteriomes protects symbiotic bacteria from host immune system.Microbiome. 2022 Sep 27;10(1):156. doi: 10.1186/s40168-022-01334-8. Microbiome. 2022. PMID: 36163269 Free PMC article.
-
The transposable element-rich genome of the cereal pest Sitophilus oryzae.BMC Biol. 2021 Nov 9;19(1):241. doi: 10.1186/s12915-021-01158-2. BMC Biol. 2021. PMID: 34749730 Free PMC article.
References
-
- Heddi A. Endosymbiosis in the weevil of genus Sitophilus: genetic, physiological and molecular interactions among associated genomes. In: Bourtzis K, Miller A, Insect Symbiosis, editors. New York: CRC Press LLC; 2003. pp. pp 67–82.
-
- Heddi A, Charles H, Khatchadourian C, Bonnot G, Nardon P. Molecular characterization of the principal symbiotic bacteria of the weevil Sitophilus oryzae: a peculiar G + C content of an endocytobiotic DNA. J Mol Evol. 1998;47:52–61. - PubMed
-
- Charles H, Heddi A, Rahbe Y. A putative insect intracellular endosymbiont stem clade, within the enterobacteriaceae, infered from phylogenetic analysis based on a heterogeneous model of DNA evolution. C R Acad Sci III. 2001;324:489–494. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

