The short-chain fatty acid receptor GPR43 is transcriptionally regulated by XBP1 in human monocytes
- PMID: 25633224
- PMCID: PMC4311239
- DOI: 10.1038/srep08134
The short-chain fatty acid receptor GPR43 is transcriptionally regulated by XBP1 in human monocytes
Abstract
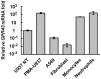
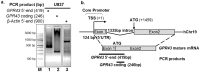
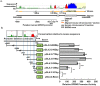
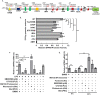
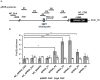
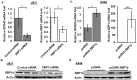
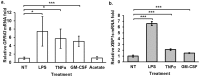
G-protein coupled receptor 43 (GPR43) recognizes short chain fatty acids and is implicated in obesity, colitis, asthma and arthritis. Here, we present the first full characterization of the GPR43 promoter and 5'-UTR. 5'-RACE of the GPR43 transcript identified the transcription start site (TSS) and a 124 bp 5'-UTR followed by a 1335 bp intron upstream of the ATG start codon. The sequence spanning -4560 to +68 bp relative to the GPR43 TSS was found to contain strong promoter activity, increasing luciferase reporter expression by >100-fold in U937 monocytes. Stepwise deletions further narrowed the putative GPR43 promoter (-451 to +68). Site-directed mutagenesis identified XBP1 as a core cis element, the mutation of which abrogated transcriptional activity. Mutations of predicted CREB, CHOP, NFAT and STAT5 binding sites, partially reduced promoter activity. ChIP assays confirmed the binding of XBP1 to the endogenous GPR43 promoter. Consistently, GPR43 expression is reduced in monocytes upon siRNA-knockdown of XBP1, while A549 cells overexpressing XBP1 displayed elevated GPR43 levels. Based on its ability to activate XBP1, we predicted and confirmed that TNFα induces GPR43 expression in human monocytes. Altogether, our findings form the basis for strategic modulation of GPR43 expression, with a view to regulate GPR43-associated diseases.
Figures
References
-
- Sina C. et al. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J. Immunol. Baltim. Md 1950 183, 7514–7522 (2009). - PubMed
-
- Kim M. H., Kang S. G., Park J. H., Yanagisawa M. & Kim C. H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145, 396–406. e1–10 (2013). - PubMed
-
- Masui R. et al. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm. Bowel Dis. 19, 2848–2856 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

