Prioritizing causal disease genes using unbiased genomic features
- PMID: 25633252
- PMCID: PMC4279789
- DOI: 10.1186/s13059-014-0534-8
Prioritizing causal disease genes using unbiased genomic features
Abstract
Background: Cardiovascular disease (CVD) is the leading cause of death in the developed world. Human genetic studies, including genome-wide sequencing and SNP-array approaches, promise to reveal disease genes and mechanisms representing new therapeutic targets. In practice, however, identification of the actual genes contributing to disease pathogenesis has lagged behind identification of associated loci, thus limiting the clinical benefits.
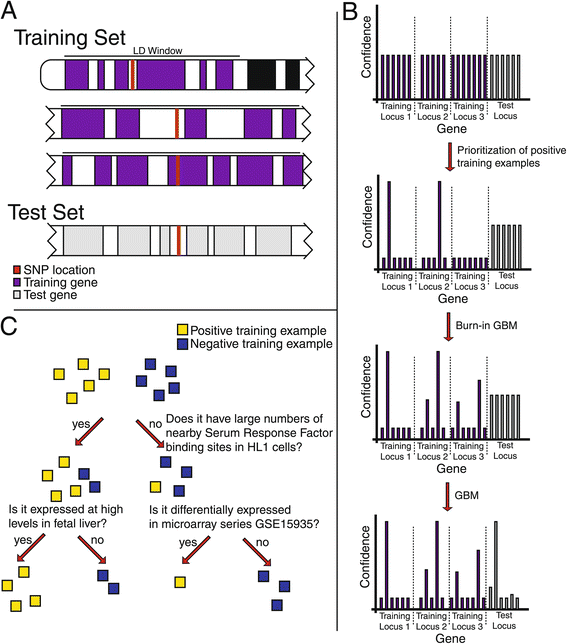
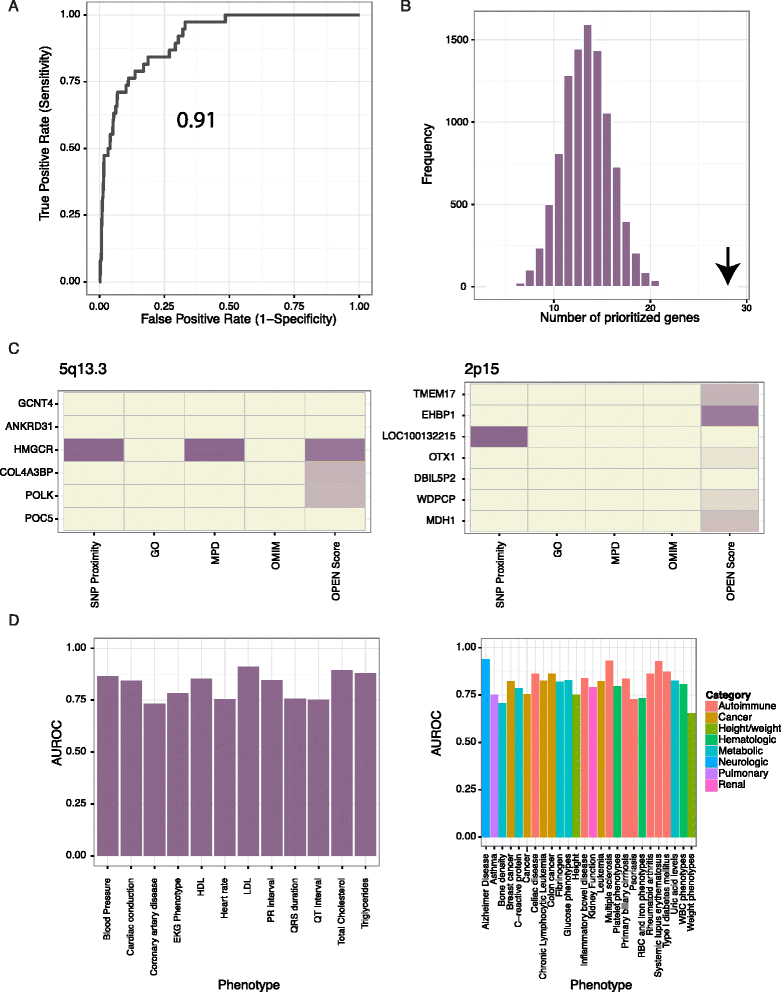
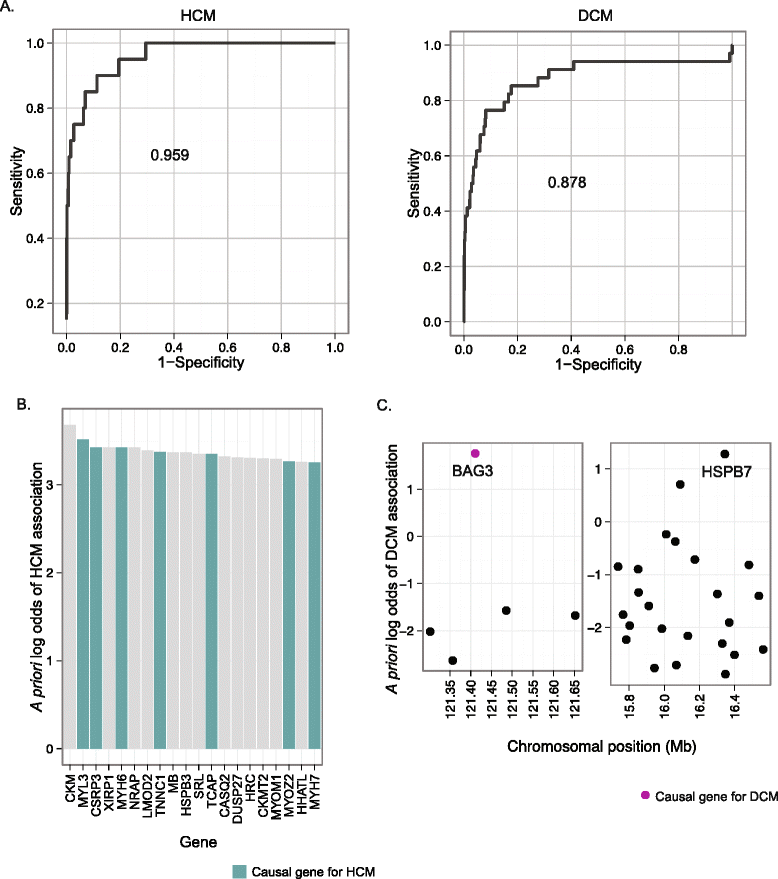
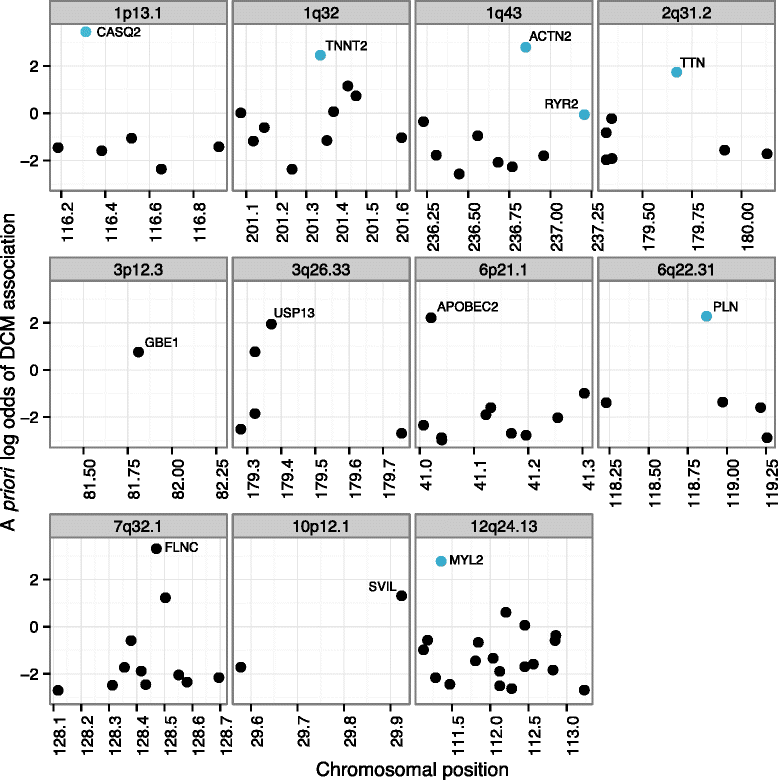
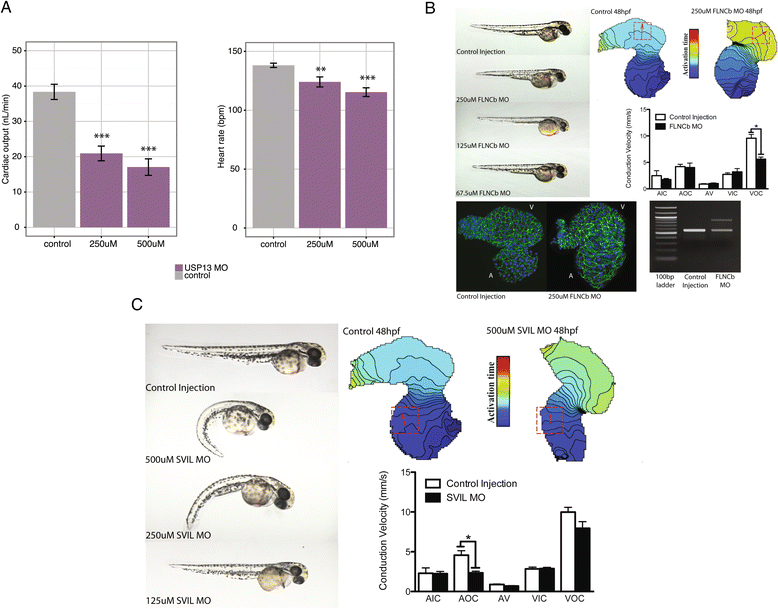
Results: To aid in localizing causal genes, we develop a machine learning approach, Objective Prioritization for Enhanced Novelty (OPEN), which quantitatively prioritizes gene-disease associations based on a diverse group of genomic features. This approach uses only unbiased predictive features and thus is not hampered by a preference towards previously well-characterized genes. We demonstrate success in identifying genetic determinants for CVD-related traits, including cholesterol levels, blood pressure, and conduction system and cardiomyopathy phenotypes. Using OPEN, we prioritize genes, including FLNC, for association with increased left ventricular diameter, which is a defining feature of a prevalent cardiovascular disorder, dilated cardiomyopathy or DCM. Using a zebrafish model, we experimentally validate FLNC and identify a novel FLNC splice-site mutation in a patient with severe DCM.
Conclusion: Our approach stands to assist interpretation of large-scale genetic studies without compromising their fundamentally unbiased nature.
Figures
References
-
- Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

