Evolutionary histories of transposable elements in the genome of the largest living marsupial carnivore, the Tasmanian devil
- PMID: 25633377
- PMCID: PMC4408412
- DOI: 10.1093/molbev/msv017
Evolutionary histories of transposable elements in the genome of the largest living marsupial carnivore, the Tasmanian devil
Abstract
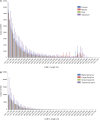
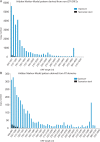
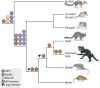
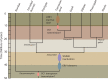
The largest living carnivorous marsupial, the Tasmanian devil (Sarcophilus harrisii), is the sole survivor of a lineage originating about 12 Ma. We set out to investigate the spectrum of transposable elements found in the Tasmanian devil genome, the first high-coverage genome of an Australian marsupial. Marsupial genomes have been shown to have the highest amount of transposable elements among vertebrates. We analyzed the horizontally transmitted DNA transposons OC1 and hAT-1_MEu in the Tasmanian devil genome. OC1 is present in all carnivorous marsupials, while having a very limited distribution among the remaining Australian marsupial orders. In contrast, hAT-1_MEu is present in all Australian marsupial orders, and has so far only been identified in a few placental mammals. We screened 158 introns for phylogenetically informative retrotransposons in the order Dasyuromorphia, and found that the youngest SINE (Short INterspersed Element), WSINE1, is no longer active in the subfamily Dasyuridae. The lack of detectable WSINE1 activity in this group may be due to a retrotransposon inactivation event approximately 30 Ma. We found that the Tasmanian devil genome contains a relatively low number of continuous full-length LINE-1 (Long INterspersed Element 1, L1) retrotransposons compared with the opossum genome. Furthermore, all L1 elements in the Tasmanian devil appeared to be nonfunctional. Hidden Markov Model approaches suggested that other potential sources of functional reverse transcriptase are absent from the genome. We discuss the issues associated with assembling long, highly similar L1 copies from short read Illumina data and describe how assembly artifacts can potentially lead to erroneous conclusions.
Keywords: DNA transposon; Sarcophilus; retrotransposon.
© The Author 2015. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures

References
-
- Birney E. Assemblies: the good, the bad, the ugly. Nat Methods. 2011;8:59–60. - PubMed
-
- Brown OJF. Tasmanian devil (Sarcophilus harrisii) extinction on the Australian mainland in the mid-Holocene: multicausality and ENSO intensification. Alcheringa. 2006;31:49–57.
-
- Cantrell MA, Grahn RA, Scott L, Wichman HA. Isolation of markers from recently transposed LINE-1 retrotransposons. Biotechniques. 2000;29:1310–1316. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

