Screening and functional pathway analysis of genes associated with pediatric allergic asthma using a DNA microarray
- PMID: 25633562
- PMCID: PMC4394950
- DOI: 10.3892/mmr.2015.3277
Screening and functional pathway analysis of genes associated with pediatric allergic asthma using a DNA microarray
Abstract
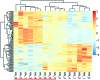
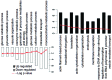
The present study aimed to identify differentially expressed genes (DEGs) associated with pediatric allergic asthma, and to analyze the functional pathways of the selected target genes, in order to explore the pathogenesis of the disease. The GSE18965 gene expression profile was downloaded from the Gene Expression Omnibus database and was preprocessed. This gene expression profile consisted of seven normal samples and nine samples from patients with pediatric allergic asthma. The DEGs between the normal and pediatric allergic asthma samples were screened using limma package in R, and the cut‑off value was set at false discovery rate <0.05 and log fold change >1. Following hierarchical clustering of the DEGs based on the expression profiles, the up‑ and downregulated genes underwent a functional enrichment analysis by topological approach (P<0.05), using the Database for Annotation, Visualization and Integrated Discovery. A total of 127 DEGs were identified between the normal and pediatric allergic asthma samples. The up‑ and downregulated genes were significantly enriched in the actin filament‑based process and the monosaccharide metabolic process, respectively. Seven downregulated DEGs (M6PR, TPP1, GLB1, NEU1, ACP2, LAMP1 and HGSNAT) were identified in the lysosomal pathway, with P=6.4x10(‑9). These results suggested that variation in lysosomal function, triggered by the seven downregulated genes, may lead to aberrant functioning of the T lymphocytes, resulting in asthma. Further research regarding the treatment of pediatric allergic asthma through targeting lysosomal function is required.
Figures

Similar articles
-
Screening of differentially expressed genes and small molecule drugs of pediatric allergic asthma with DNA microarray.Eur Rev Med Pharmacol Sci. 2012 Dec;16(14):1961-6. Eur Rev Med Pharmacol Sci. 2012. PMID: 23242723
-
Protein-protein interaction network analysis and identifying regulation microRNAs in asthmatic children.Allergol Immunopathol (Madr). 2015 Nov-Dec;43(6):584-92. doi: 10.1016/j.aller.2015.01.008. Epub 2015 May 12. Allergol Immunopathol (Madr). 2015. PMID: 25979194
-
[Comparative analysis of the role of CD4(+) and CD8(+) T cells in severe asthma development].Mol Biol (Mosk). 2015 May-Jun;49(3):482-90. doi: 10.7868/S0026898415030180. Mol Biol (Mosk). 2015. PMID: 26107902 Russian.
-
Identification of the differentially expressed genes associated with familial combined hyperlipidemia using bioinformatics analysis.Mol Med Rep. 2015 Jun;11(6):4032-8. doi: 10.3892/mmr.2015.3263. Epub 2015 Jan 27. Mol Med Rep. 2015. PMID: 25625967 Free PMC article.
-
Uncovering potential key genes associated with the pathogenesis of asthma: A microarray analysis of asthma-relevant tissues.Allergol Immunopathol (Madr). 2017 Mar-Apr;45(2):152-159. doi: 10.1016/j.aller.2016.08.007. Epub 2016 Nov 11. Allergol Immunopathol (Madr). 2017. PMID: 27842724
Cited by
-
Expression and Regulation of Thymic Stromal Lymphopoietin and Thymic Stromal Lymphopoietin Receptor Heterocomplex in the Innate-Adaptive Immunity of Pediatric Asthma.Int J Mol Sci. 2018 Apr 18;19(4):1231. doi: 10.3390/ijms19041231. Int J Mol Sci. 2018. PMID: 29670037 Free PMC article. Review.
-
Chromatin accessibility profiling of Treg cells in acute urticaria.Epigenetics. 2025 Dec;20(1):2503126. doi: 10.1080/15592294.2025.2503126. Epub 2025 May 12. Epigenetics. 2025. PMID: 40355834 Free PMC article.
-
Targeting protein glycosylation to regulate inflammation in the respiratory tract: novel diagnostic and therapeutic candidates for chronic respiratory diseases.Front Immunol. 2023 May 15;14:1168023. doi: 10.3389/fimmu.2023.1168023. eCollection 2023. Front Immunol. 2023. PMID: 37256139 Free PMC article. Review.
-
The Exploitation of the Glycosylation Pattern in Asthma: How We Alter Ancestral Pathways to Develop New Treatments.Biomolecules. 2024 Apr 24;14(5):513. doi: 10.3390/biom14050513. Biomolecules. 2024. PMID: 38785919 Free PMC article. Review.
-
Methylomic markers of persistent childhood asthma: a longitudinal study of asthma-discordant monozygotic twins.Clin Epigenetics. 2015 Dec 18;7:130. doi: 10.1186/s13148-015-0163-4. eCollection 2015. Clin Epigenetics. 2015. PMID: 26691723 Free PMC article.
References
-
- Mannino DM, Homa DM, Akinbami LJ, Moorman JE, Gwynn C, Redd SC. Surveillance for asthma - United States, 1980–1999. MMWR Surveill Summ. 2002;51:1–13. - PubMed
-
- Wang XQ, Wang XM, Zhou TF, Dong LQ. Screening of differentially expressed genes and small molecule drugs of pediatric allergic asthma with DNA microarray. Eur Rev Med Pharmacol Sci. 2012;16:1961–1966. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

