Direct Detection and Quantification of Bacterial Genes Associated with Inflammation in DNA Isolated from Stool
- PMID: 25635239
- PMCID: PMC4307837
- DOI: 10.4236/aim.2014.415117
Direct Detection and Quantification of Bacterial Genes Associated with Inflammation in DNA Isolated from Stool
Abstract
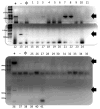
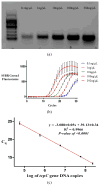
Although predominantly associated with health benefits, the gut microbiota has also been shown to harbor genes that promote inflammation. In this work, we report a method for the direct detection and quantification of these pro-inflammatory bacterial genes by PCR and qPCR in DNA extracted from human stool samples. PCR reactions were performed to detect (i) the pks island genes, (ii) tcpC, which is present in some strains of Escherichia coli and (iii) gelE presented in some strains of Enterococcus faecalis. Additionally, we screened for the presence of the following genes encoding cyclomodulins that disrupted mammalian cell division: (iv) cdt (which encodes the cytolethal distending toxin) and (v) cnf-1 (which encodes the cytotoxic necrotizing factor-1). Our results show that 20% of the samples (N = 41) tested positive for detectable amounts of pks island genes, whereas 10% of individuals were positive for tcpC or gelE and only one individual was found to harbor the cnf-1 gene. Of the 13 individuals that were positive for at least one of the pro-inflammatory genes, 5 were found to harbor more than one. A quantitative version of the assay, which used real-time PCR, revealed the pro-inflammatory genes to be in high copy numbers: up to 1.3 million copies per mg of feces for the pks island genes. Direct detection of specific genes in stool could prove useful toward screening for the presence of pro-inflammatory bacterial genes in individuals with inflammatory bowel diseases or colorectal cancer.
Keywords: Gut Inflammation; Microbial Biomarkers; Microbiota; PCR; Stool Samples.
Figures

References
-
- Balter M. Taking Stock of the Human Microbiome and Disease. Science. 2012;336:1246–1247. http://dx.doi.org/10.1126/science.336.6086.1246. - DOI - PubMed
-
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the Human Intestinal Microbial Flora. Science. 2005;308:1635–1638. http://dx.doi.org/10.1126/science.1110591. - DOI - PMC - PubMed
-
- Blaser M, Bork P, Fraser C, Knight R, Wang J. The Microbiome Explored: Recent Insights and Future Challenges. Nature Reviews Microbiology. 2013;11:213–217. http://dx.doi.org/10.1038/nrmicro2973. - DOI - PubMed
-
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J, Hansen T, Le Paslier D, Linneberg A, Nielsen H, Bjorn, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Dusko Ehrlich S, Wang J. A Human Gut Microbial Gene Catalogue Established by Metagenomic Sequencing. Nature. 2010;464:59–65. http://dx.doi.org/10.1038/nature08821. - DOI - PMC - PubMed
-
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics Analysis Reveals Large Effects of Gut Microflora on Mammalian Blood Metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:3698–3703. http://dx.doi.org/10.1073/pnas.0812874106. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
