A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes
- PMID: 25635642
- PMCID: PMC4438322
- DOI: 10.1038/ismej.2014.220
A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes
Abstract
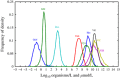
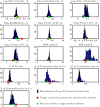
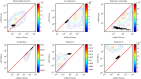
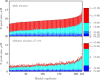
Viral lysis of microbial hosts releases organic matter that can then be assimilated by nontargeted microorganisms. Quantitative estimates of virus-mediated recycling of carbon in marine waters, first established in the late 1990s, were originally extrapolated from marine host and virus densities, host carbon content and inferred viral lysis rates. Yet, these estimates did not explicitly incorporate the cascade of complex feedbacks associated with virus-mediated lysis. To evaluate the role of viruses in shaping community structure and ecosystem functioning, we extend dynamic multitrophic ecosystem models to include a virus component, specifically parameterized for processes taking place in the ocean euphotic zone. Crucially, we are able to solve this model analytically, facilitating evaluation of model behavior under many alternative parameterizations. Analyses reveal that the addition of a virus component promotes the emergence of complex communities. In addition, biomass partitioning of the emergent multitrophic community is consistent with well-established empirical norms in the surface oceans. At steady state, ecosystem fluxes can be probed to characterize the effects that viruses have when compared with putative marine surface ecosystems without viruses. The model suggests that ecosystems with viruses will have (1) increased organic matter recycling, (2) reduced transfer to higher trophic levels and (3) increased net primary productivity. These model findings support hypotheses that viruses can have significant stimulatory effects across whole-ecosystem scales. We suggest that existing efforts to predict carbon and nutrient cycling without considering virus effects are likely to miss essential features of marine food webs that regulate global biogeochemical cycles.
Figures


References
-
- Armstrong RA, McGehee R. Competitive exclusion. Am Nat. 1980;115:151–170.
-
- Barber RT, Marra J, Bidigare RC, Codispoti LA, Halpern D, Johnson Z, et al. Primary productivity and its regulation in the Arabian Sea during 1995. Deep Sea Res Part II: Top Stud Oceanogr. 2001;48:1127–1172.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

