YAP regulates S-phase entry in endothelial cells
- PMID: 25635998
- PMCID: PMC4312014
- DOI: 10.1371/journal.pone.0117522
YAP regulates S-phase entry in endothelial cells
Abstract
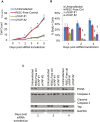
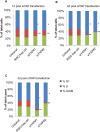
The Hippo pathway regulates cell proliferation and apoptosis through the Yes-associated protein (YAP) transcriptional activator. YAP has a well-described role in promoting cell proliferation and survival, but the precise mechanisms and transcriptional targets that underlie these properties are still unclear and likely context-dependent. We found, using siRNA-mediated knockdown, that YAP is required for proliferation in endothelial cells but not HeLa cells. Specifically, YAP is required for S-phase entry and its absence causes cells to accumulate in G1. Microarray analysis suggests that YAP mediates this effect by regulating the transcription of genes involved in the assembly and/or firing of replication origins and homologous recombination of DNA. These findings thus provide insight into the molecular mechanisms by which YAP regulates cell cycle progression.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

