NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven quiescence programmes
- PMID: 25636082
- PMCID: PMC4313575
- DOI: 10.1038/ncomms7170
NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven quiescence programmes
Abstract
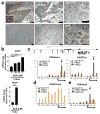
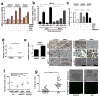
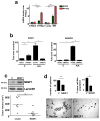
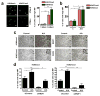
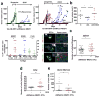
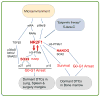
Metastases can originate from disseminated tumour cells (DTCs), which may be dormant for years before reactivation. Here we find that the orphan nuclear receptor NR2F1 is epigenetically upregulated in experimental head and neck squamous cell carcinoma (HNSCC) dormancy models and in DTCs from prostate cancer patients carrying dormant disease for 7-18 years. NR2F1-dependent dormancy is recapitulated by a co-treatment with the DNA-demethylating agent 5-Aza-C and retinoic acid across various cancer types. NR2F1-induced quiescence is dependent on SOX9, RARβ and CDK inhibitors. Intriguingly, NR2F1 induces global chromatin repression and the pluripotency gene NANOG, which contributes to dormancy of DTCs in the bone marrow. When NR2F1 is blocked in vivo, growth arrest or survival of dormant DTCs is interrupted in different organs. We conclude that NR2F1 is a critical node in dormancy induction and maintenance by integrating epigenetic programmes of quiescence and survival in DTCs.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

